5CGB
 
 | | Crystal structure of FimH in complex with heptyl alpha-D-septanoside | | Descriptor: | (6R)-1,6-anhydro-2-O-heptyl-6-(hydroxymethyl)-D-galactitol, Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Preston, R.P, Zihlmann, P, Fiege, B, Sager, C.P, Vannam, R, Rabbani, S, Zalewski, A, Maier, T, Ernst, B, Peczuh, M. | | Deposit date: | 2015-07-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The price of flexibility - a case study on septanoses as pyranose mimetics.
Chem Sci, 9, 2018
|
|
6DR0
 
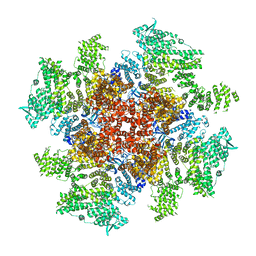 | | Class 5 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7L3H
 
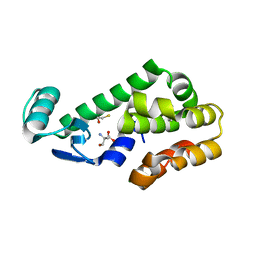 | | T4 Lysozyme L99A - ethylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
4XZ4
 
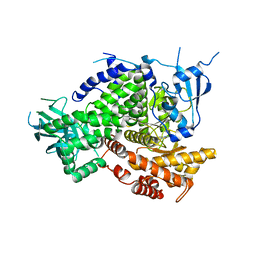 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
8R49
 
 | |
5BWD
 
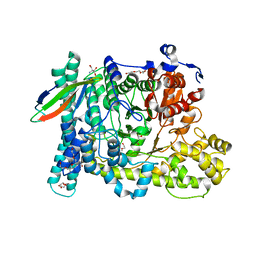 | | Benzylsuccinate alpha-gamma bound to fumarate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FUMARIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-06-07 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-bound Structures of Benzylsuccinate Synthase Reveal How Toluene Is Activated in Anaerobic Hydrocarbon Degradation.
J.Biol.Chem., 290, 2015
|
|
7L3K
 
 | | T4 Lysozyme L99A - benzylacetate - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
4MJS
 
 | | crystal structure of a PB1 complex | | Descriptor: | 1,2-ETHANEDIOL, Protein kinase C zeta type, Sequestosome-1 | | Authors: | Ren, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2013-09-04 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the homotypic PB1-PB1 complex between PKC zeta and p62
Sci China Life Sci, 57, 2014
|
|
7L3C
 
 | | T4 Lysozyme L99A - o-xylene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
4XX5
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
7L3I
 
 | | T4 Lysozyme L99A - propylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endolysin, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
1PMN
 
 | | Crystal structure of JNK3 in complex with an imidazole-pyrimidine inhibitor | | Descriptor: | CYCLOPROPYL-{4-[5-(3,4-DICHLOROPHENYL)-2-[(1-METHYL)-PIPERIDIN]-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
5S74
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 Nsp3 macrodomain
To Be Published
|
|
7KLQ
 
 | |
3E4A
 
 | | Human IDE-inhibitor complex at 2.6 angstrom resolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, HYDROXAMATE PEPTIDE II1, ... | | Authors: | Malito, E, Leissring, M.A, Choi, S, Cuny, G.D, Tang, W.J. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin.
Plos One, 5, 2010
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 16 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
4Y3C
 
 | | I304V 3D polymerase mutant of EMCV | | Descriptor: | 3D polymerase, CHLORIDE ION, GLYCEROL | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
5H14
 
 | | EED in complex with an allosteric PRC2 inhibitor EED666 | | Descriptor: | 2-[3-(3,5-dimethylpyrazol-1-yl)-4-nitro-phenyl]-3,4-dihydro-1H-isoquinoline, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
6PIU
 
 | |
4Y3D
 
 | |
6PIY
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-2 (NR02-61) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
3L9H
 
 | | X-ray structure of mitotic kinesin-5 (KSP, KIF11, Eg5)in complex with the hexahydro-2H-pyrano[3,2-c]quinoline EMD 534085 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-{[(2R,4aS,5R,10bS)-5-phenyl-9-(trifluoromethyl)-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinolin-2-yl]methyl}urea, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11 | | Authors: | Knoechel, T. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery and optimization of hexahydro-2H-pyrano[3,2-c]quinolines (HHPQs) as potent and selective inhibitors of the mitotic kinesin-5.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3HBX
 
 | | Crystal structure of GAD1 from Arabidopsis thaliana | | Descriptor: | Glutamate decarboxylase 1 | | Authors: | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
6GCC
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor in complex with dextran-sulfate | | Descriptor: | 3,4-di-O-sulfo-alpha-D-glucopyranose-(1-6)-3,4-di-O-sulfo-alpha-D-altropyranose-(1-6)-1,2,3,4-tetra-O-sulfo-alpha-D-allopyranose, Glutathione transferase Xi 3 mutant C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-04-17 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
6SB7
 
 | | Human Carbonic Anhydrase II in complex with fragment. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-chloranylthiophene-2-sulfonamide, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-19 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
