2ZTS
 
 | |
2ZMV
 
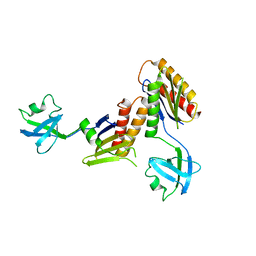 | | Crystal structure of Synbindin | | Descriptor: | Trafficking protein particle complex subunit 4 | | Authors: | Fan, F. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human synbindin reveals two conformations of longin domain
Biochem.Biophys.Res.Commun., 378, 2009
|
|
3QPG
 
 | |
2ZSO
 
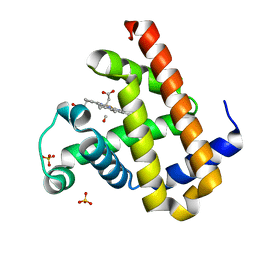 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser on [450 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3QN6
 
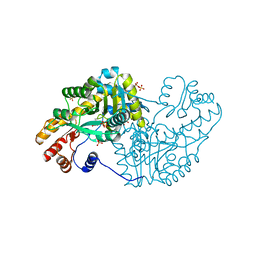 | |
3BRD
 
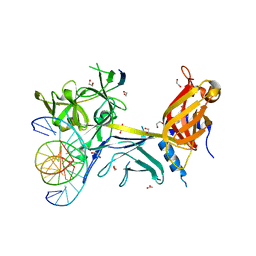 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
3BRW
 
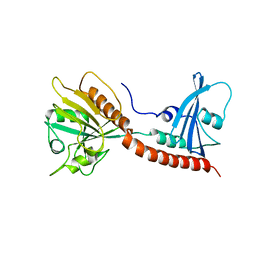 | | Structure of the Rap-RapGAP complex | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Scrima, A, Thomas, C, Deaconescu, D, Wittinghofer, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues
Embo J., 27, 2008
|
|
3BUN
 
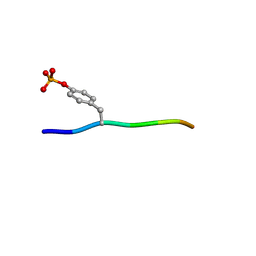 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty4 | | Descriptor: | 13-meric peptide from Protein sprouty homolog 4, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BW9
 
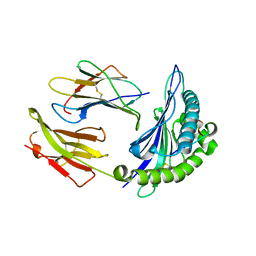 | | Crystal Structure of HLA B*3508 in complex with a HCMV 12-mer peptide from the pp65 protein | | Descriptor: | Beta-2-microglobulin, CPS peptide from 65 kDa lower matrix phosphoprotein, HLA class I histocompatibility antigen, ... | | Authors: | Wynn, K.K, Marland, Z, Cooper, L, Silins, S.L, Gras, S, Archbold, J.K, Tynan, F.E, Miles, J.J, McCluskey, J, Burrows, S.R, Rossjohn, J, Khanna, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection
Blood, 111, 2008
|
|
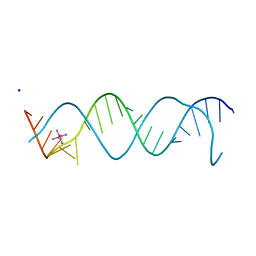
3BNT
 
 | |
3BOL
 
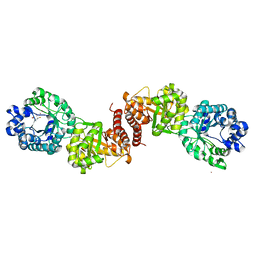 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7B9V
 
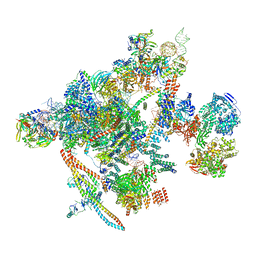 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | Descriptor: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
3T05
 
 | |
3BUI
 
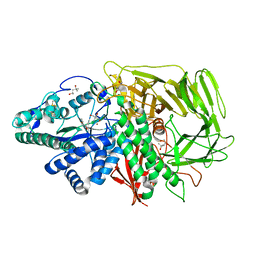 | | Golgi mannosidase II D204A catalytic nucleophile mutant complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-mannosidase 2, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-01-02 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the substrate specificity of Golgi alpha-mannosidase II by use of synthetic oligosaccharides and a catalytic nucleophile mutant.
J.Am.Chem.Soc., 130, 2008
|
|
3C2G
 
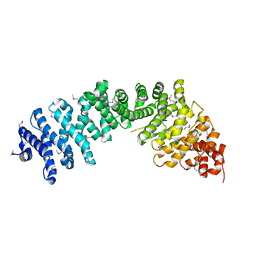 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
3BVT
 
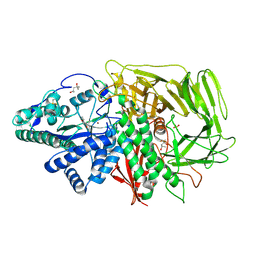 | | GOLGI MANNOSIDASE II D204A catalytic nucleophile mutant complex with Methyl (alpha-D-mannopyranosyl)-(1->3)-S-alpha-D-mannopyranoside | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-01-07 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the substrate specificity of Golgi alpha-mannosidase II by use of synthetic oligosaccharides and a catalytic nucleophile mutant.
J.Am.Chem.Soc., 130, 2008
|
|
3T6B
 
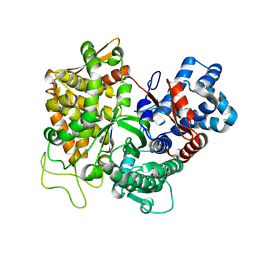 | |
3BKL
 
 | | Testis ACE co-crystal structure with ketone ACE inhibitor kAW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Watermeyer, J.M, Kroger, W.L, O'Neill, H.G, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2007-12-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Probing the basis of domain-dependent inhibition using novel ketone inhibitors of Angiotensin-converting enzyme
Biochemistry, 47, 2008
|
|
3BLQ
 
 | | Crystal Structure of Human CDK9/cyclinT1 in Complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Cell division protein kinase 9, ... | | Authors: | Baumli, S, Lolli, G, Lowe, E.D, Johnson, L.N. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation
Embo J., 27, 2008
|
|
3BXZ
 
 | | Crystal structure of the isolated DEAD motor domains from Escherichia coli SecA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase subunit secA, ... | | Authors: | Nithianantham, S, Namjoshi, S, Shilton, B.H. | | Deposit date: | 2008-01-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Analysis of the isolated SecA DEAD motor suggests a mechanism for chemical-mechanical coupling.
J.Mol.Biol., 383, 2008
|
|
3BN9
 
 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
3BZL
 
 | | Crystal structural of native EscU C-terminal domain | | Descriptor: | EscU, FORMIC ACID, SODIUM ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BP4
 
 | | The high resolution crystal structure of HLA-B*2705 in complex with a Cathepsin A signal sequence peptide pCatA | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
3TC1
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | Descriptor: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | Authors: | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
3C2E
 
 | |
