1A7E
 
 | |
9E9I
 
 | | Crystal Structure of human KRAS G12C covalently bound to nopinone-derived naphthol compound 21 | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(4P,6R,8R)-3-fluoro-4-(3-hydroxynaphthalen-1-yl)-7,7-dimethyl-5,6,7,8-tetrahydro-6,8-methanoquinolin-2-yl]-2,6-diazaspiro[3.4]octan-2-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2024-11-08 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Identification of Structurally Novel KRAS G12C Inhibitors through Covalent DNA-Encoded Library Screening.
J.Med.Chem., 68, 2025
|
|
8F0G
 
 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
9BIG
 
 | | Stat6 bound to degrader AK-1690 | | Descriptor: | Signal transducer and activator of transcription 6, [(2-{[(2S)-1-{(2S,4S)-4-[(7-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}hept-6-yn-1-yl)oxy]-2-[(2R)-2-phenylmorpholine-4-carbonyl]pyrrolidin-1-yl}-3,3-dimethyl-1-oxobutan-2-yl]carbamoyl}-1-benzothiophen-5-yl)di(fluoro)methyl]phosphonic acid | | Authors: | Mallik, L, Stuckey, J.A. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Discovery of AK-1690: A Potent and Highly Selective STAT6 PROTAC Degrader.
J.Med.Chem., 68, 2025
|
|
7LTQ
 
 | |
5KWW
 
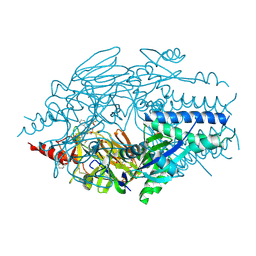 | | Crystal Structure of Inhibitor JNJ-53718678 In Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[5-chloranyl-1-(3-methylsulfonylpropyl)indol-2-yl]methyl]-1-[2,2,2-tris(fluoranyl)ethyl]imidazo[4,5-c]pyridin-2-one, ... | | Authors: | McLellan, J.S, Battles, M.B, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2016-07-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Therapeutic efficacy of a respiratory syncytial virus fusion inhibitor.
Nat Commun, 8, 2017
|
|
3MPX
 
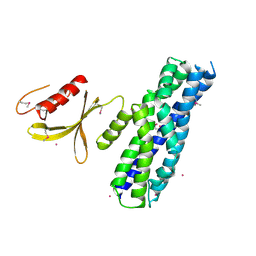 | | Crystal structure of the DH and PH-1 domains of human FGD5 | | Descriptor: | FYVE, RhoGEF and PH domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Nedyalkova, L, Tong, Y, Tempel, W, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the DH and PH-1 domains of human FGD5
TO BE PUBLISHED
|
|
6GYO
 
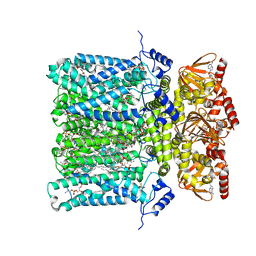 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel in complex with cAMP | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
9EU1
 
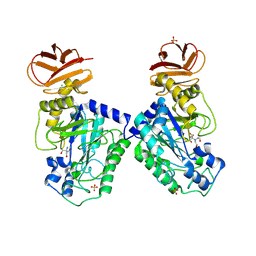 | | GH29A alpha-L-fucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-fucosidase, ... | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
6BXS
 
 | |
5YPW
 
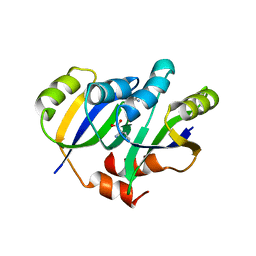 | | Crystal structure of IlvN.Val-1b | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5HCT
 
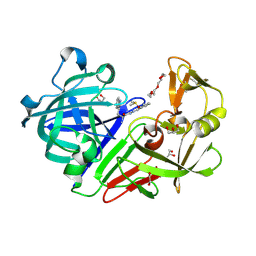 | | Endothiapepsin in complex with biacylhydrazone | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-N'-{3-[(E)-{2-[(2S)-2-amino-3-(1H-indol-3-yl)propanoyl]hydrazinylidene}methyl]benzylidene}-3-(1H-indol-2-yl)propanehydrazide (non-preferred name), ACETATE ION, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-01-04 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin: Fragment-Based Drug Design Facilitated by Dynamic Combinatorial Chemistry.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
7TBE
 
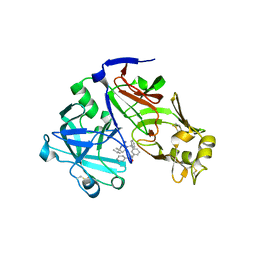 | | Crystal structure of Plasmepsin X from Plasmodium vivax in complex with WM4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(R)-[(2E,4S)-2-imino-4-methyl-6-oxo-4-(propan-2-yl)-1,3-diazinan-1-yl](phenyl)methyl]-N-[(1S)-1-phenylethyl]benzamide, Plasmepsin X, ... | | Authors: | Hodder, A.N, Christensen, J.B, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
9BS5
 
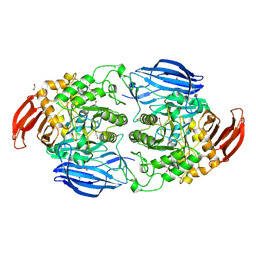 | | Bacteroides ovatus GH97C Sus | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | Authors: | Brown, H.A, Koropatkin, N.M. | | Deposit date: | 2024-05-13 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Acarbose impairs gut Bacteroides growth by targeting intracellular glucosidases.
Mbio, 15, 2024
|
|
5CV1
 
 | | C. elegans PGL-1 Dimerization Domain | | Descriptor: | P granule abnormality protein 1 | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6S2W
 
 | | Structure of S. pombe Erh1, a protein important for meiotic mRNA decay in mitosis and meiosis progression. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Hazra, D, Graille, M. | | Deposit date: | 2019-06-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Formation of S. pombe Erh1 homodimer mediates gametogenic gene silencing and meiosis progression.
Sci Rep, 10, 2020
|
|
7TBD
 
 | | Crystal structure of Plasmepsin X from Plasmodium vivax in complex with WM382 | | Descriptor: | (4R)-4-[(2E)-4,4-diethyl-2-imino-6-oxo-1,3-diazinan-1-yl]-N-[(4S)-2,2-dimethyl-3,4-dihydro-2H-1-benzopyran-4-yl]-3,4-dihydro-2H-1-benzopyran-6-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hodder, A.N, Christensen, J.B, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
9BMK
 
 | | GH5_4 endo-beta(1,3/1,4)-glucanase E331A from Segatella copri in complex with cellotriose | | Descriptor: | Cellulase (Glycosyl hydrolase family 5), SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cordeiro, R.L, Golisch, B, Brumer, H, Van Petegem, F. | | Deposit date: | 2024-05-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The molecular basis of cereal mixed-linkage beta-glucan utilization by the human gut bacterium Segatella copri.
J.Biol.Chem., 300, 2024
|
|
8ER7
 
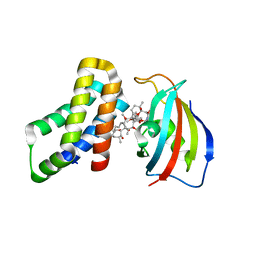 | | FKBP12-FRB in Complex with Compound 12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-9,27-dihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-5,10,21-trimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
8G7Q
 
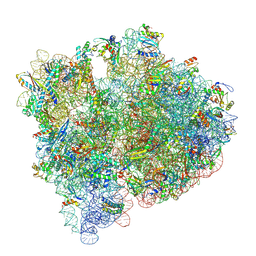 | |
7JMJ
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 37 - State 5 (S5) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
8BXW
 
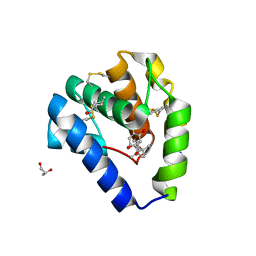 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Carvacrol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-BUTANOL, 2-methyl-5-propan-2-yl-phenol, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
6ZSG
 
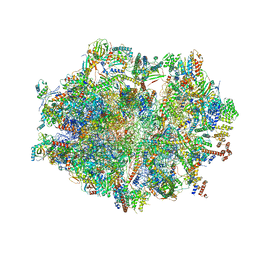 | | Human mitochondrial ribosome in complex with mRNA, A-site tRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
9N9X
 
 | | Structure of HPK1 with C5 bound at its active site | | Descriptor: | (1R,2S)-2-cyanocyclopentyl [(6P)-8-amino-7-fluoro-6-(8-methyl-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)isoquinolin-3-yl]carbamate, 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase kinase 1, ... | | Authors: | Kiefer, J.R, Wang, W, Wu, P, Walters, B.T, Dou, Y. | | Deposit date: | 2025-02-11 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
