8BGF
 
 | |
3VHT
 
 | | Crystal structure of GFP-Wrnip1 UBZ domain fusion protein in complex with ubiquitin | | Descriptor: | Green fluorescent protein, Green fluorescent protein,ATPase WRNIP1, Ubiquitin, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2011-09-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 2016
|
|
7ABH
 
 | | Human pre-Bact-2 spliceosome (SF3b/U2 snRNP portion) | | Descriptor: | Cell division cycle 5-like protein, DNA/RNA-binding protein KIN17, MINX M3 pre-mRNA, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
3VHS
 
 | |
6ZLX
 
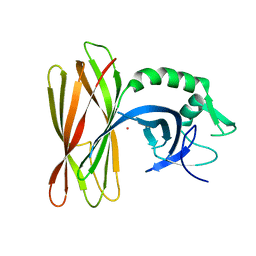 | | The structure of the ClpX-associated factor PDIP38 | | Descriptor: | PLATINUM (II) ION, Polymerase delta-interacting protein 2 | | Authors: | Zeth, K, Dougan, D. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | PDIP38 is a novel adaptor-like modulator of the mitochondrial AAA+ protease CLPXP
To Be Published
|
|
4PXM
 
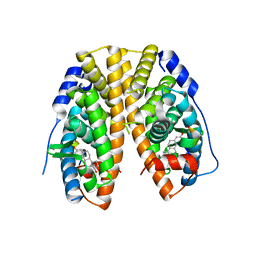 | |
6EG1
 
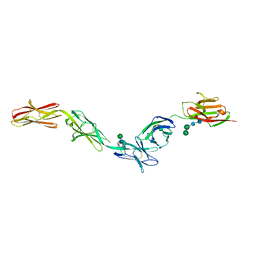 | | Crystal structure of Dpr2 Ig1-Ig2 in complex with DIP-Theta Ig1-Ig3 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cosmanescu, F, Patel, S, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6EFZ
 
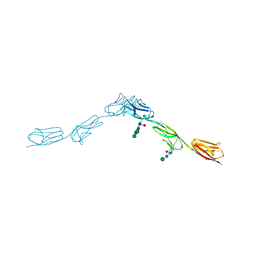 | | Crystal Structure of DIP-Theta Ig1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6ERF
 
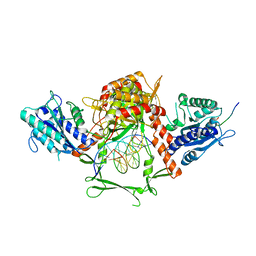 | | Complex of APLF factor and Ku heterodimer bound to DNA | | Descriptor: | Aprataxin and PNK-like factor, DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat.Struct.Mol.Biol., 25, 2018
|
|
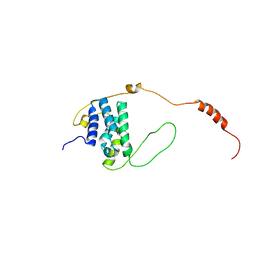
6EMR
 
 | |
6EMP
 
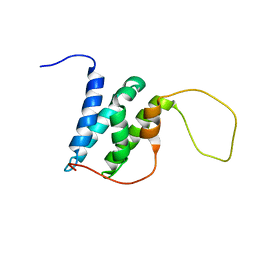 | | Solution structure of the LEDGF/p75 IBD - POGZ (aa 1370-1404) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Pogo transposable element with ZNF domain | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMO
 
 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 1-32) complex | | Descriptor: | PC4 and SFRS1-interacting protein,LEDGF/p75 IBD-JPO2 M1 | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMQ
 
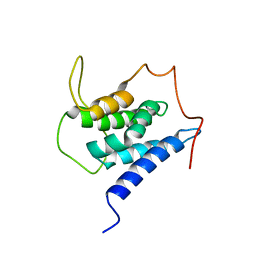 | | Solution structure of the LEDGF/p75 IBD - MLL1 (aa 111-160) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Histone-lysine N-methyltransferase 2A | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6V7O
 
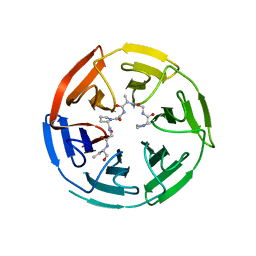 | |
5NG2
 
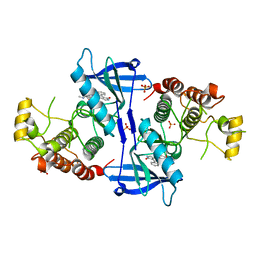 | | Structure of RIP2K(D146N) with bound Staurosporine | | Descriptor: | PHOSPHATE ION, Receptor-interacting serine/threonine-protein kinase 2, STAUROSPORINE | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
3WH0
 
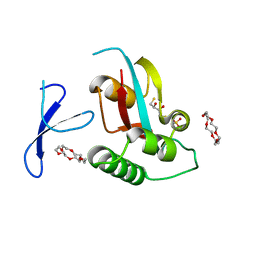 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7BQX
 
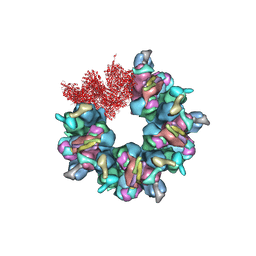 | | Epstein-Barr virus, C5 portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
1ZCN
 
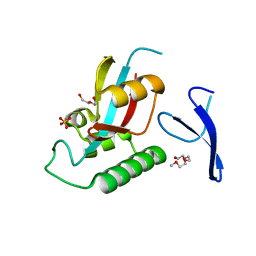 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4ROJ
 
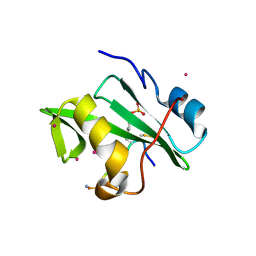 | | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide | | Descriptor: | Guanine nucleotide exchange factor VAV2, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide
TO BE PUBLISHED
|
|
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
3ZEH
 
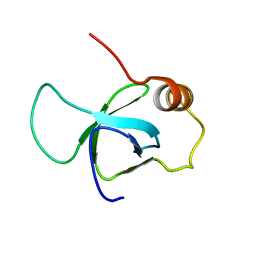 | |
8E3H
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E5P
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E6W
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, lambda-tR1 rut RNA, Mg-ADP-BeF3, and NusG; Rho part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E70
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, dC75 rut mimic RNA, Mg-ADP-BeF3, and NusG; Rho hexamer part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
