6QYL
 
 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
3EKY
 
 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
9GCR
 
 | | Human Butyrylcholinesterase in complex with N1,N1-dimethyl-N2-(6-(naphthalen-1-yl)-5-(pyridin-4-yl)pyridazin-3-yl)ethane-1,2-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2024-08-02 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Human Butyrylcholinesterase in complex with N1,N1-dimethyl-N2-(6-(naphthalen-1-yl)-5-(pyridin-4-yl)pyridazin-3-yl)ethane-1,2-diamine
To Be Published
|
|
9DIE
 
 | | Structure of phospholipase D BetaIB1i from Sicarius terrosus venom, H47N mutant bound to product and substrate sphingolipids at 1.85 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-aminoethyl (2S,3R,4E)-2-dodecanamido-3-hydroxyheptadec-4-en-1-yl hydrogen (S)-phosphate, Dermonecrotic toxin StSicTox-betaIB1i, ... | | Authors: | Sundman, A.K, Montfort, W.R, Binford, G.J, Cordes, M.H. | | Deposit date: | 2024-09-05 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of phospholipase D BetaIB1i from Sicarius terrosus venom, H47N mutant bound to product and substrate sphingolipids at 1.85 A resolution
To Be Published
|
|
4ZAD
 
 | | Structure of C. dubliensis Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
3UYT
 
 | | crystal structure of ck1d with PF670462 from P1 crystal form | | Descriptor: | 4-[1-cyclohexyl-4-(4-fluorophenyl)-1H-imidazol-5-yl]pyrimidin-2-amine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2011-12-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
6QZ7
 
 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4Z34
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4NZ7
 
 | | Steroid receptor RNA Activator (SRA) modification by the human Pseudouridine Synthase 1 (hPus1p): RNA binding, activity, and atomic model | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TRIETHYLENE GLYCOL, tRNA pseudouridine synthase A, ... | | Authors: | Huet, T, Thore, S. | | Deposit date: | 2013-12-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Steroid Receptor RNA Activator (SRA) Modification by the Human Pseudouridine Synthase 1 (hPus1p): RNA Binding, Activity, and Atomic Model
Plos One, 9, 2014
|
|
7UGN
 
 | |
5CUG
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 4-Acetamidobenzoic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, PARA ACETAMIDO BENZOIC ACID | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 4-Acetamidobenzoic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
4RN3
 
 | |
7UGO
 
 | |
7UGQ
 
 | |
4DSD
 
 | |
4ZPE
 
 | |
8D0W
 
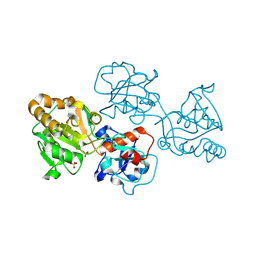 | | Human FUT9 bound to H-Type 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
5GZ7
 
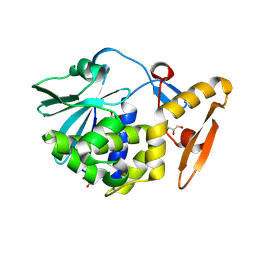 | | Crystal Structure of the complex of Ribosome Inactivating Protein with 1,2-ethanediol at 1.95 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tiwari, P, Pandey, S.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-09-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the complex of Ribosome Inactivating Protein with 1,2-ethanediol at 1.95 Angstrom resolution.
To Be Published
|
|
2VPN
 
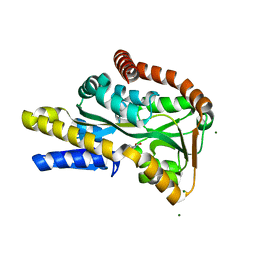 | | High-resolution structure of the periplasmic ectoine-binding protein from TeaABC TRAP-transporter of Halomonas elongata | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, MAGNESIUM ION, PERIPLASMIC SUBSTRATE BINDING PROTEIN | | Authors: | Kuhlmann, S.I, Terwisscha van Scheltinga, A.C, Bienert, R, Kunte, H.J, Ziegler, C. | | Deposit date: | 2008-03-03 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 A Structure of the Ectoine Binding Protein Teaa of the Osmoregulated Trap-Transporter Teaabc from Halomonas Elongata.
Biochemistry, 47, 2008
|
|
7VHY
 
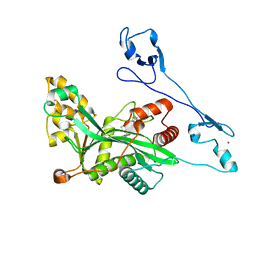 | | Crystal structure of EP300 HAT domain in complex with compound (+)-3 | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(6R)-6-(1H-indazol-4-ylmethyl)-1,4-oxazepan-4-yl]-[1-(4-methoxyphenyl)cyclopentyl]methanone | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
8CQL
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-N-((S)-1-(5-Fluoro-2-methoxy-4-(4-methylthiazol-5-yl)phenyl)ethyl)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamide (Compound 33) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[5-fluoranyl-2-methoxy-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
8CQK
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 30) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[2-methyl-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
7W2H
 
 | |
4ZPG
 
 | |
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
