5L0R
 
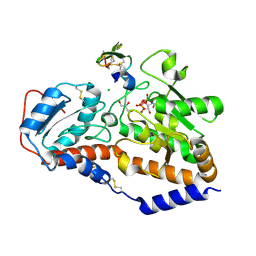 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
9J5V
 
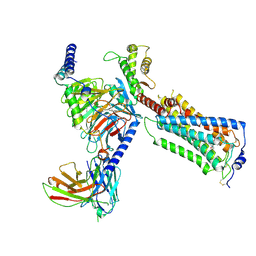 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to CpY | | Descriptor: | CpY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2024-08-13 | | Release date: | 2024-11-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural mechanisms of potent lysophosphatidic acid receptor 1 activation by nonlipid basic agonists.
Commun Biol, 7, 2024
|
|
6GN6
 
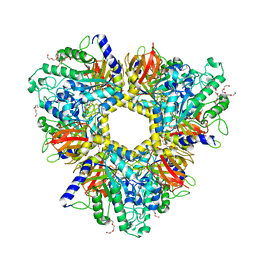 | | Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Kovalova, T, Koval, T, Lipovova, P, Dohnalek, J. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active site complementation and hexameric arrangement in the GH family 29; a structure-function study of alpha-l-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus.
Glycobiology, 29, 2019
|
|
6L17
 
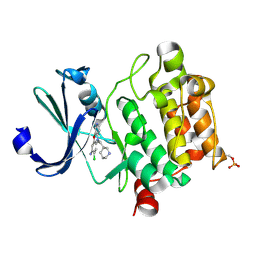 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
7S8V
 
 | |
6UUL
 
 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6R3O
 
 | | Aspergillus niger ferric acid decarboxylase (Fdc) L439G variant in complex with prFMN (purified in the radical form) and phenylpropiolic acid | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2019-03-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Atomic description of an enzyme reaction dependent on reversible 1,3-dipolar cycloaddition
to be published
|
|
6FS1
 
 | | MCL1 in complex with an indole acid ligand | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Kasmirski, S, Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6L01
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 2-[3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]phenyl]ethanoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6KZO
 
 | | membrane protein | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6R56
 
 | | Crystal structure of PPEP-1(K101E/E184K) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
5CU8
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2-Amino-6-chlorobenzothiazole (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-1,3-benzothiazol-2-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2-Amino-6-chlorobenzothiazole (SGC - Diamond I04-1 fragment screening)
To be published
|
|
5CUE
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N,1-dimethyl-1H-pyrazole-4-carboxamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening)
To be published
|
|
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
6KZV
 
 | |
6R53
 
 | | Crystal structure of PPEP-1(K101R) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6CNX
 
 | | Crystal Structure of the Human vaccinia-related kinase 1 (VRK1) bound to an N-propynyl-N-isopentyl-dihydropteridin inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-8-(3-methylbutyl)-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, (7S)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-8-(3-methylbutyl)-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Vaccinia-Related Kinase 1 (VRK1) bound to a N-propynyl-N-isopentil-dihydropteridine inhibitor
To Be Published
|
|
5ODH
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 3.5 minutes | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
6BSL
 
 | | BMP1 complexed with a reverse hydroxymate - compound 22 | | Descriptor: | 1,2-ETHANEDIOL, Bone morphogenetic protein 1, N-(2-ethoxy-4-{5-[({[(2R)-2-{(1R)-1-[formyl(hydroxy)amino]propyl}heptanoyl]amino}methyl)carbamoyl]furan-2-yl}benzene-1-carbonyl)-L-aspartic acid, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
7CHT
 
 | | Crystal structure of TTK kinase domain in complex with compound 30 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(oxan-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK, MAGNESIUM ION | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Ko, E.H, Choi, H.G, Son, J.B, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
3AUA
 
 | | Crystal structure of the quaternary complex-2 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
6KZZ
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
7CHN
 
 | | Crystal structure of TTK kinase domain in complex with compound 9 | | Descriptor: | 4-(cyclohexylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
6C7I
 
 | | Crystal structure of human phosphodiesterase 2A with 1-(2-chloro-5-methoxy-phenyl)-N-isobutyl-4-methyl-[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide | | Descriptor: | 1-(2-chloro-5-methoxyphenyl)-4-methyl-N-(2-methylpropyl)[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2018-01-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J. Med. Chem., 61, 2018
|
|
