7AD2
 
 | |
1KRH
 
 | | X-ray Structure of Benzoate Dioxygenase Reductase | | Descriptor: | Benzoate 1,2-Dioxygenase Reductase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Karlsson, A, Beharry, Z.M, Eby, D.M, Coulter, E.D, Niedle, E.L, Kurtz Jr, D.M, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1.
J.Mol.Biol., 318, 2002
|
|
7A9K
 
 | |
7YWP
 
 | | Closed conformation of Oligopeptidase B from Serratia proteomaculans with covalently bound TCK | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inhibitor-Bound Bacterial Oligopeptidase B in the Closed State: Similarity and Difference between Protozoan and Bacterial Enzymes.
Int J Mol Sci, 24, 2023
|
|
1KLQ
 
 | | The Mad2 Spindle Checkpoint Protein Undergoes Similar Major Conformational Changes upon Binding to Either Mad1 or Cdc20 | | Descriptor: | MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A, Mad2-binding peptide | | Authors: | Luo, X, Tang, Z, Rizo, J, Yu, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20.
Mol.Cell, 9, 2002
|
|
1KLZ
 
 | |
1KS0
 
 | | The First Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | Matrix Metalloproteinase 2 | | Authors: | Gehrmann, M, Briknarova, K, Banyai, L, Patthy, L, Llinas, M. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The col-1 module of human matrix metalloproteinase-2 (MMP-2): structural/functional relatedness between gelatin-binding fibronectin type II modules and lysine-binding kringle domains.
Biol.Chem., 383, 2002
|
|
1KT5
 
 | | Crystal structure of bovine holo-RBP at pH 4.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KTH
 
 | |
7A6V
 
 | |
1KM6
 
 | |
1KMG
 
 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
1KPP
 
 | | Structure of the Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Rich, R.L, Myszka, D.G, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-01-02 | | Release date: | 2002-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functional interactions of the Tsg101 UEV domain.
EMBO J., 21, 2002
|
|
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KTO
 
 | |
1KU2
 
 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment Containing Regions 1.2 to 3.1 | | Descriptor: | SULFATE ION, sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1KV6
 
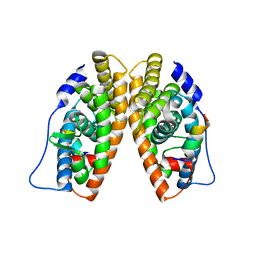 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
1KQU
 
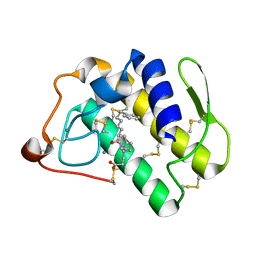 | | Human phospholipase A2 complexed with a substrate anologue | | Descriptor: | 6-PHENYL-4(R)-(7-PHENYL-HEPTANOYLAMINO)-HEXANOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Tyndall, J.D, Martin, J.L. | | Deposit date: | 2002-01-07 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D-Tyrosine as a chiral precusor to potent inhibitors of human nonpancreatic secretory phospholipase A2 (IIa) with antiinflammatory activity.
Chembiochem, 4, 2003
|
|
9CJA
 
 | |
7AEZ
 
 | |
1KR2
 
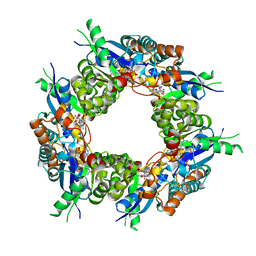 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
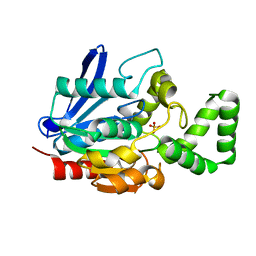
7A6G
 
 | |
7YWZ
 
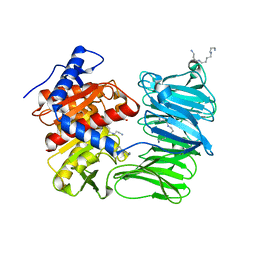 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution | | Descriptor: | GLYCEROL, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution
To Be Published
|
|
1KVE
 
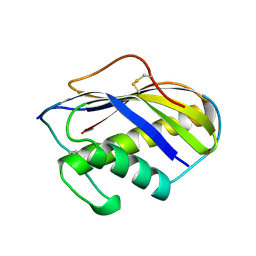 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
6VYP
 
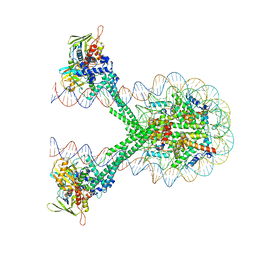 | | Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate | | Descriptor: | DNA (191-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A type 1, ... | | Authors: | Kim, S, Zhu, J, Eek, P, Yennawar, N, Song, T. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.99 Å) | | Cite: | Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.
Mol.Cell, 78, 2020
|
|
