5X7O
 
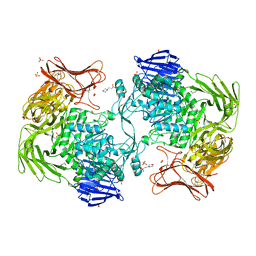 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Suzuki, N, Kishine, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
1APT
 
 | |
4PWD
 
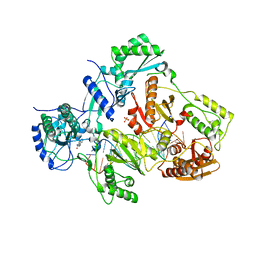 | | Crystal structure of HIV-1 reverse transcriptase in complex with bulge-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*U)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-19 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
5VJE
 
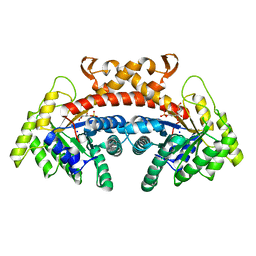 | | Class II fructose-1,6-bisphosphate aldolase of Escherichia coli with D-glucitol 1,6-bisphosphate | | Descriptor: | D-Glucitol-1,6-bisphosphate, Fructose-bisphosphate aldolase class 2, SODIUM ION | | Authors: | Coincon, M, Sygusch, J. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
3IS9
 
 | | Crystal structure of the HIV-1 reverse transcriptase (RT) in complex with the alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate). | | Descriptor: | Reverse transcriptase, Reverse transcriptase/ribonuclease H, dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate) | | Authors: | Ho, W.C, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2009-08-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic study of a novel subnanomolar inhibitor provides insight on the binding interactions of alkenyldiarylmethanes with human immunodeficiency virus-1 reverse transcriptase.
J.Med.Chem., 52, 2009
|
|
5CLM
 
 | | 1,4-Oxazine BACE1 inhibitors | | Descriptor: | Beta-secretase 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rombouts, F, Tresadern, G, Delgado, O, Martinez Lamenca, C, Van Gool, M, Garcia-Molina, A, Alonso De Diego, S, Oehlrich, D, Prokopcova, H, Alonso, J.M, Austin, N, Borghys, H, Van Brandt, S, Surkyn, M, De Cleyn, M, Vos, A, Alexander, R, MacDonald, G, Moechars, D, Trabanco, A, Gijsen, H. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 1,4-Oxazine beta-Secretase 1 (BACE1) Inhibitors: From Hit Generation to Orally Bioavailable Brain Penetrant Leads.
J.Med.Chem., 58, 2015
|
|
5VQU
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide (JLJ683), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Petrova, Z.O, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4TPP
 
 | | 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors | | Descriptor: | 1-[4-(3-{[1-(quinolin-2-yl)azetidin-3-yl]oxy}quinoxalin-2-yl)piperidin-1-yl]ethanone, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-06-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility.
Bioorg.Med.Chem., 22, 2014
|
|
5UBT
 
 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A 7-(3-(PIPERAZIN-1-YL)PHENYL)PYRROLO[2,1-F][1,2,4] TRIAZIN-4-AMINE DERIVIATINE | | Descriptor: | 1-[4-(3-{4-amino-5-[1-(oxan-4-yl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)piperazin-1-yl]ethan-1-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Sack, J.S. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of 7-(3-(piperazin-1-yl)phenyl)pyrrolo[2,1-f][1,2,4]triazin-4-amine derivatives as highly potent and selective PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5CPQ
 
 | | Disproportionating enzyme 1 from Arabidopsis - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
3KLE
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5UBR
 
 | | CRYSTAL STRUCTURE OF PI3K ALPHA IN COMPLEX WITH A 7-(3-(PIPERAZIN-1-YL)PHENYL)PYRROLO[2,1-F][1,2,4] TRIAZIN-4-AMINE DERIVIATINE | | Descriptor: | 1-[4-(3-{4-amino-5-[1-(oxan-4-yl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)piperazin-1-yl]ethan-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Sack, J.S. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of 7-(3-(piperazin-1-yl)phenyl)pyrrolo[2,1-f][1,2,4]triazin-4-amine derivatives as highly potent and selective PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5VQQ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide (JLJ683), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQY
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQV
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7D0T
 
 | |
6AEF
 
 | | PapA2 acyl transferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Trehalose-2-sulfate acyltransferase PapA2, ... | | Authors: | Chaudhary, S, Rao, V, Panchal, V. | | Deposit date: | 2018-08-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A novel mutation alters the stability of PapA2 resulting in the complete abrogation of sulfolipids in clinical mycobacterial strains.
Faseb Bioadv, 1, 2019
|
|
4YDI
 
 | | Crystal structure of broad and potently neutralizing VRC01-class antibody Z258-VRC27.01, isolated from human donor Z258, in complex with HIV-1 gp120 from clade A strain Q23.17 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.452 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4RWY
 
 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC131 in complex with HIV-1 clade B YU2 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
5VQT
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | 2-chloro-N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Czyzyk, D.J, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6CKT
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Legionella pneumophila Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Legionella pneumophila Philadelphia 1
to be published
|
|
6G5T
 
 | | Myoglobin H64V/V68A in the resting state, 1.5 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
4Q0B
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with gap-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-04-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
2WIP
 
 | | STRUCTURE OF CDK2-CYCLIN A COMPLEXED WITH 8-ANILINO-1-METHYL-4,5-DIHYDRO- 1H-PYRAZOLO[4,3-H] QUINAZOLINE-3-CARBOXYLIC ACID | | Descriptor: | 1-methyl-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxylic acid, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
2XEZ
 
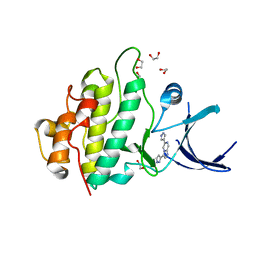 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 6-(1H-PYRAZOL-3-YL)-3-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, vanMontfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | Deposit date: | 2010-05-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
