8XPP
 
 | |
3H5X
 
 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
3H5Y
 
 | | Norovirus polymerase+primer/template+CTP complex at 6 mM MnCl2 | | Descriptor: | 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
2CKW
 
 | | The 2.3 A resolution structure of the Sapporo virus RNA dependant RNA polymerase. | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Fullerton, S.W.B, Tucker, P.A, Rohayem, J, Coutard, B, Gebhardt, J, Gorbalenya, A, Canard, B. | | Deposit date: | 2006-04-24 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Sapovirus RNA-Dependent RNA Polymerase.
J.Virol., 81, 2007
|
|
2B43
 
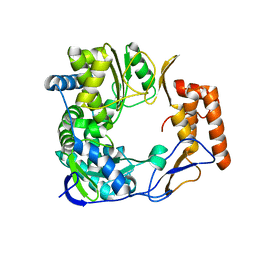 | |
5Y3D
 
 | |
6S2L
 
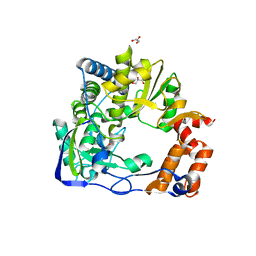 | |
3UQS
 
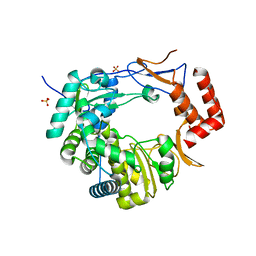 | |
5ZIT
 
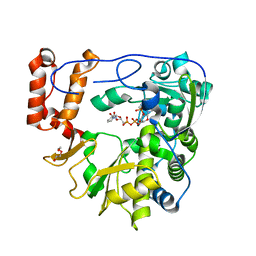 | | Crystal structure of human Enterovirus D68 RdRp in complex with NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RdRp | | Authors: | Wang, M.L, Li, L, Chen, Y.P, Jiang, H, Zhang, Y, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 2020
|
|
5ZIU
 
 | | Crystal structure of human Entervirus D68 RdRp | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RdRp | | Authors: | Wang, M.L, Zhang, Y, Chen, Y.P, Lu, D.R, Jiang, H, Chen, Y.J, Li, L, Zhang, C.H, Shi, Q.L, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Crystal structure of human Entervirus D68 RdRp in complex with NADPH
To Be Published
|
|
6R1I
 
 | | Structure of porcine Aichi virus polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
6QWT
 
 | | Sicinivirus 3Dpol RNA dependent RNA polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
3UR0
 
 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
3UPF
 
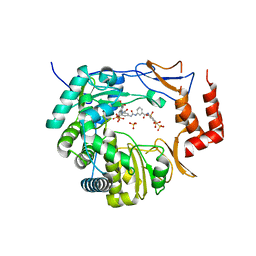 | | Crystal structure of murine norovirus RNA-dependent RNA polymerase bound to NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
6KWR
 
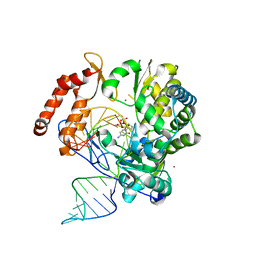 | |
6KWQ
 
 | |
6L4R
 
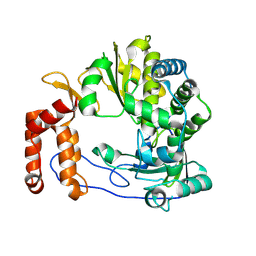 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
2ILZ
 
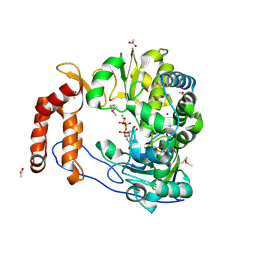 | |
2IJF
 
 | |
2IM0
 
 | |
6LSE
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C3S6A/C3S6B form) | | Descriptor: | Genome polyprotein, PYROPHOSPHATE 2-, RNA (35-MER), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
3BSN
 
 | |
3BSO
 
 | |
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3CDW
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
