6QII
 
 | | Xenon derivatization of the F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QGT
 
 | | The carbon monoxide inhibition of F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QGR
 
 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8POU
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.65 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POV
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.92 A Resolution. | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
7ODH
 
 | |
7ODG
 
 | |
8POW
 
 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.61 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
4CI0
 
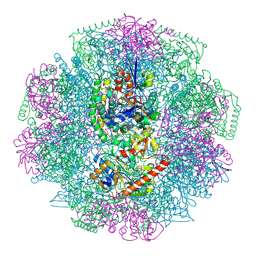 | | Electron cryo-microscopy of F420-reducing NiFe hydrogenase Frh | | Descriptor: | F420-REDUCING HYDROGENASE, SUBUNIT ALPHA, SUBUNIT BETA, ... | | Authors: | Allegretti, M, Mills, D.J, McMullan, G, Kuehlbrandt, W, Vonck, J. | | Deposit date: | 2013-12-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Atomic Model of the F420-Reducing [Nife] Hydrogenase by Electron Cryo-Electron Microscopy Using a Direct Electron Detector.
Elife, 3, 2014
|
|
3ZEA
 
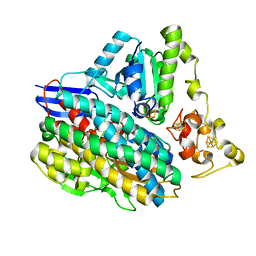 | | 3D structure of the NiFeSe hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.82 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
4C3O
 
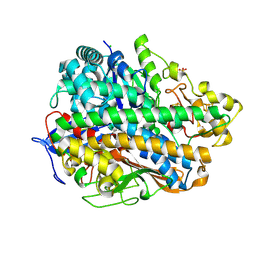 | | Structure and function of an oxygen tolerant NiFe hydrogenase from Salmonella | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Bowman, L, Flanagan, L, Fyfe, P.K, Parkin, A, Hunter, W.N, Sargent, F. | | Deposit date: | 2013-08-26 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How the Structure of the Large Subunit Controls Function in an Oxygen-Tolerant [Nife]-Hydrogenase.
Biochem.J., 458, 2014
|
|
7VXQ
 
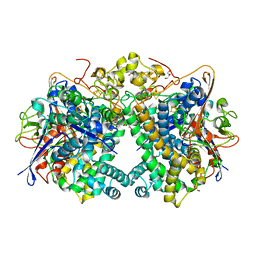 | | The Carbon Monoxide Complex of [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 | | Descriptor: | CARBON MONOXIDE, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Higuchi, K, Imanishi, T, Higuchi, Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and spectroscopic characterization of CO inhibition of [NiFe]-hydrogenase from Citrobacter sp. S-77.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7UUR
 
 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
5A4F
 
 | | The mechanism of Hydrogen Activation by NiFe-hydrogenases. | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5AA5
 
 | | Actinobacterial-type NiFe-hydrogenase from Ralstonia eutropha H16 at 2.85 Angstrom resolution | | Descriptor: | IRON/SULFUR CLUSTER, MALONIC ACID, NIFE-HYDROGENASE LARGE SUBUNIT, ... | | Authors: | Schaefer, C, Bommer, M, Hennig, S, Jeoung, J.H, Dobbek, H, Lenz, O. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structure of an Actinobacterial-Type [Nife]-Hydrogenase Reveals Insight Into O2-Tolerant H2 Oxidation.
Structure, 24, 2016
|
|
5A4I
 
 | | The mechanism of Hydrogen activation by NiFE-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.C, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5A4M
 
 | | Mechanism of Hydrogen activation by NiFe-hydrogenases | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sergent, F, Carr, S.B, Philips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
6RTP
 
 | |
6RUC
 
 | |
6RU9
 
 | |
1FRF
 
 | | CRYSTAL STRUCTURE OF THE NI-FE HYDROGENASE FROM DESULFOVIBRIO FRUCTOSOVORANS | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Montet, Y, Volbeda, A, Piras, C, Hatchikian, E.C, Frey, M, Fontecilla, J.C. | | Deposit date: | 1998-07-21 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3Fe-4S] to [4Fe-4S] cluster conversion in Desulfovibrio fructosovorans [NiFe] hydrogenase by site-directed mutagenesis
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1E3D
 
 | | [NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), FE3-S4 CLUSTER, ... | | Authors: | Matias, P.M, Soares, C.M, Saraiva, L.M, Coelho, R, Morais, J, Le Gall, J, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with the Tetrahaem Cytochrome C3.
J.Biol.Inorg.Chem., 6, 2001
|
|
6SYO
 
 | | Hydrogenase-2 variant R479K - As Isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
