8PR4
 
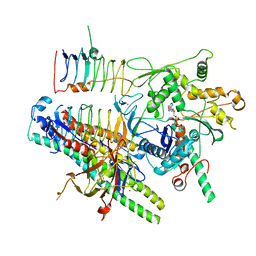 | | Dynactin pointed end bound to JIP3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Arp11, C-Jun-amino-terminal kinase-interacting protein 3, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PXW
 
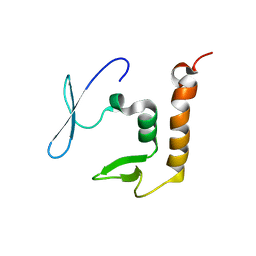 | |
5F0J
 
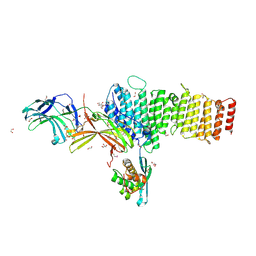 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
8QEY
 
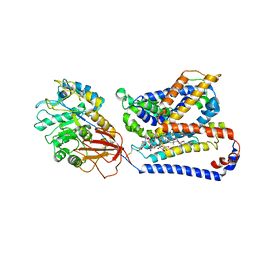 | | Structure of human Asc1/CD98hc heteromeric amino acid transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Martinez-Molledo, M, Rullo-Tubau, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2023-09-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and mechanisms of transport of human Asc1/CD98hc amino acid transporter.
Nat Commun, 15, 2024
|
|
8Q7V
 
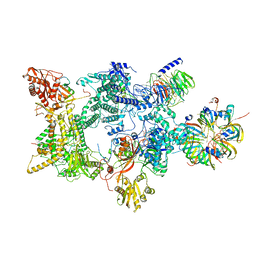 | | Structure of the recycling U5 snRNP bound to chaperones CD2BP2 and TSSC4 (State 1) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7Q
 
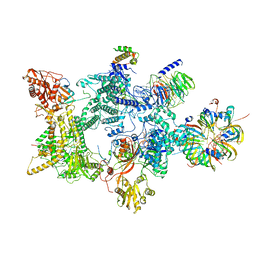 | | Structure of the recycling U5 snRNP bound to chaperones CD2BP2 and TSSC4 (State 2) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7X
 
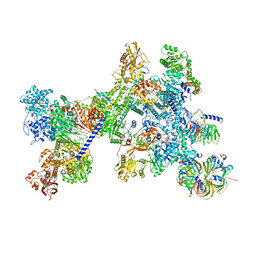 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 4) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7W
 
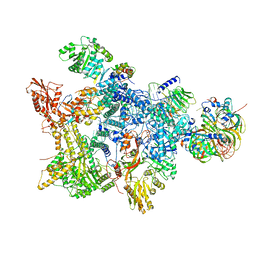 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 3) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5F0L
 
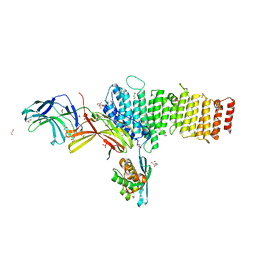 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5F2R
 
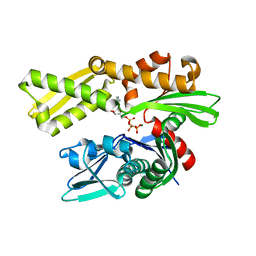 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with AMP-PCP | | Descriptor: | 78 kDa glucose-regulated protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-12-02 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F74
 
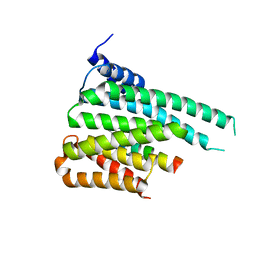 | | Crystal structure of ChREBP:14-3-3 complex bound with AMP | | Descriptor: | 14-3-3 protein beta/alpha, ADENOSINE MONOPHOSPHATE, Carbohydrate-responsive element-binding protein | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Metabolite Regulation of Nuclear Localization of Carbohydrate-response Element-binding Protein (ChREBP): ROLE OF AMP AS AN ALLOSTERIC INHIBITOR.
J.Biol.Chem., 291, 2016
|
|
5EXW
 
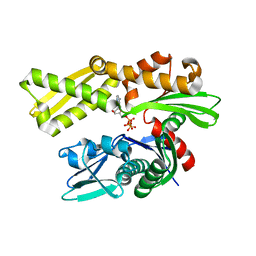 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ATP | | Descriptor: | 7-deazaadenosine-5'-triphosphate, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EW9
 
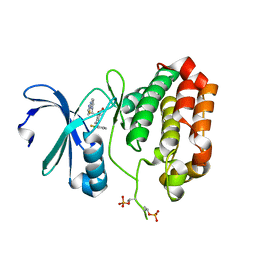 | | Crystal Structure of Aurora A Kinase Domain Bound to MK-5108 | | Descriptor: | 4-(3-chloranyl-2-fluoranyl-phenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid, Aurora kinase A | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | A Cell Biologist's Field Guide to Aurora Kinase Inhibitors.
Front Oncol, 5, 2015
|
|
8SB6
 
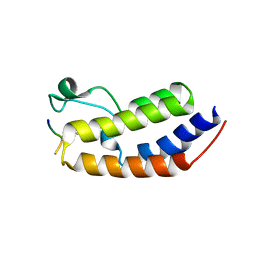 | | Structure of human BRD2-BD1 bound to a histone H4 acetyl-methyllysine peptide | | Descriptor: | Bromodomain containing 2, Histone H4 | | Authors: | Connor, L.J, Ekundayo, B.E, Lu-Culligan, W.J, Simon, M.D, Bleichert, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622, 2023
|
|
5F1X
 
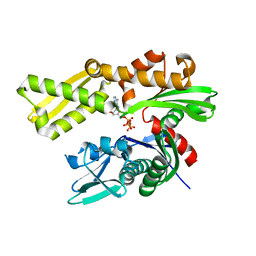 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F62
 
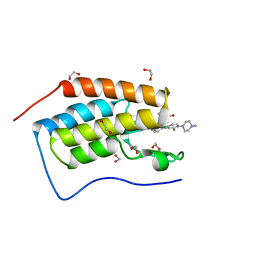 | | Crystal structure of the first bromodomain of human BRD4 in complex with MA4-022-2 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[2-chloranyl-5-[[2-[[3-fluoranyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-5-methyl-pyrimidin-4-yl]amino]phenyl]-2-methyl-propane-2-sulfonamide | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
5F4X
 
 | |
5EY4
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F66
 
 | |
5F0X
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ADP and inorganic phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 78 kDa glucose-regulated protein, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F5Z
 
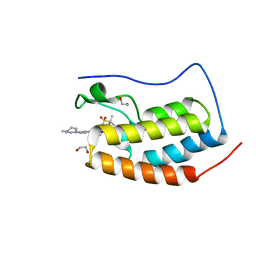 | | Crystal structure of the first bromodomain of human BRD4 in complex with MA2-014 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-~{N}-[3-[[5-methyl-2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]amino]phenyl]propane-2-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
5EX5
 
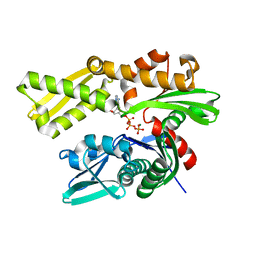 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ADP and inorganic phosphate | | Descriptor: | 7-deazaadenosine-5'-diphosphate, 78 kDa glucose-regulated protein, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F0M
 
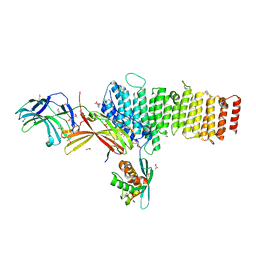 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5F60
 
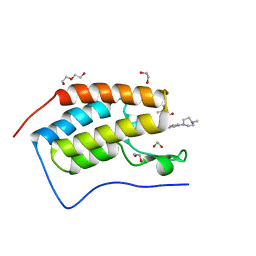 | | Crystal structure of the first bromodomain of human BRD4 in complex with SG3-014 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
8RC0
 
 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
