1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
7BIA
 
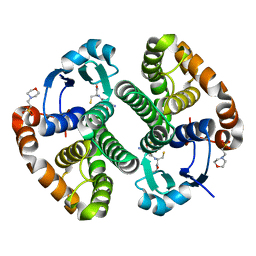 | | Crystal structure of human GSTP1 bound to iberin | | Descriptor: | 1-isothiocyanato-3-methylsulfinyl-propane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, ... | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Role of human salivary enzymes in bitter taste perception.
Food Chem, 386, 2022
|
|
5A61
 
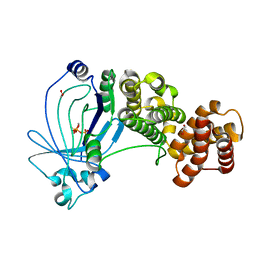 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two manganese ions. | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
1XKU
 
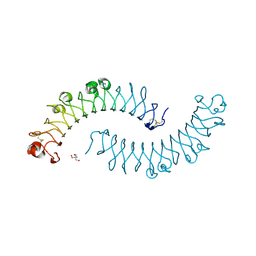 | | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Decorin | | Authors: | Scott, P.G, McEwan, P.A, Dodd, C.M, Bergmann, E.M, Bishop, P.N, Bella, J. | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8EML
 
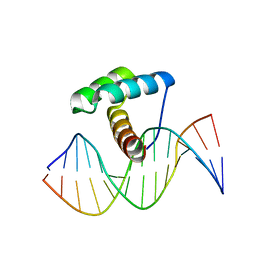 | | Crystal Structure of Gsx2 Homeodomain in Complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*GP*CP*TP*AP*AP*TP*TP*AP*AP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*TP*TP*AP*AP*TP*TP*AP*GP*CP*TP*C)-3'), GS homeobox 2, ... | | Authors: | Webb, J.A, Kovall, R.A. | | Deposit date: | 2022-09-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Cooperative Gsx2-DNA binding requires DNA bending and a novel Gsx2 homeodomain interface.
Nucleic Acids Res., 52, 2024
|
|
1YFD
 
 | | Crystal structure of the Y122H mutant of ribonucleotide reductase R2 protein from E. coli | | Descriptor: | MERCURY (II) ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Kolberg, M, Logan, D.T, Bleifuss, G, Poetsch, S, Sjoeberg, B.M, Graeslund, A, Lubitz, W, Lassmann, G, Lendzian, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies
J.Biol.Chem., 280, 2005
|
|
1FTT
 
 | | THYROID TRANSCRIPTION FACTOR 1 HOMEODOMAIN (RATTUS NORVEGICUS) | | Descriptor: | THYROID TRANSCRIPTION FACTOR 1 HOMEODOMAIN | | Authors: | Fogolari, F, Esposito, G, Damante, G, Formisano, S, Di Lauro, R, Viglino, P. | | Deposit date: | 1995-10-03 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the solution structure of the homeodomain of rat thyroid transcription factor 1 by 1H-NMR spectroscopy and restrained molecular mechanics.
Eur.J.Biochem., 241, 1996
|
|
1FTZ
 
 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
3TVD
 
 | | Crystal Structure of Mouse RhoA-GTP complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Swaminathan, K, Pal, K, Jobichen, C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Crystal structure of mouse RhoA:GTPgammaS complex in a centered lattice.
J.Struct.Funct.Genom., 13, 2012
|
|
2LD5
 
 | |
2L7Z
 
 | | NMR Structure of A13 homedomain | | Descriptor: | Homeobox protein Hox-A13 | | Authors: | Ames, J. | | Deposit date: | 2010-12-27 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence specific DNA binding and protein dimerization of HOXA13.
Plos One, 6, 2011
|
|
117E
 
 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
8PRK
 
 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
7AW9
 
 | | CCAAT-binding complex and HapX bound to Aspergillus fumigatus cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
7AW7
 
 | | CCAAT-binding complex and HapX bound to Aspergillus nidulans cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
3RSE
 
 | | Structural and biochemical characterization of two binding sites for nucleation promoting factor WASp-VCA on Arp2/3 complex | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Pollard, T.D, Jurgenson, C.T, Ti, S, Nolen, B.J. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LSP
 
 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
6FBW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-II with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
1YSO
 
 | | YEAST CU, ZN SUPEROXIDE DISMUTASE WITH THE REDUCED BRIDGE BROKEN | | Descriptor: | COPPER (I) ION, YEAST CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Crane, B.R, Tsang, J, Tainer, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
2JCW
 
 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (I) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-21 | | Release date: | 1999-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
2LP0
 
 | |
2LFB
 
 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | LFB1/HNF1 TRANSCRIPTION FACTOR | | Authors: | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | Deposit date: | 1996-12-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
2RNQ
 
 | |
3HX0
 
 | | ternary complex of L277A, H511A, R514 mutant pol lambda bound to a 2 nucleotide gapped DNA substrate with a scrunched dA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', 5'-D(*CP*GP*GP*CP*AP*AP*AP*TP*AP*CP*TP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Havener, J.M, Perera, L, Krahn, J.M, Pedersen, L.C, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2009-06-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Scrunching During DNA Repair Synthesis
To be Published
|
|
4I2B
 
 | | Ternary complex of mouse TdT with ssDNA and AMPcPP | | Descriptor: | 5'-D(P*AP*AP*AP*AP*A)-3', DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA nucleotidylexotransferase, ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Intermediates along the Catalytic Cycle of Terminal Deoxynucleotidyltransferase: Dynamical Aspects of the Two-Metal Ion Mechanism.
J.Mol.Biol., 425, 2013
|
|
