6SOM
 
 | | Metal free structure of SynFtn variant D137A | | Descriptor: | CHLORIDE ION, Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2019-08-29 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Routes of iron entry into, and exit from, the catalytic ferroxidase sites of the prokaryotic ferritin SynFtn.
Dalton Trans, 49, 2020
|
|
6ZRP
 
 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, De Luca, F, Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S, Doquier, J.D. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6SOQ
 
 | |
5LDK
 
 | |
6ZRH
 
 | | Crystal structure of OXA-10loop24 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6SIY
 
 | | PaaK family AMP-ligase with AMP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5LDP
 
 | |
5LDZ
 
 | | Quadruple space group ambiguity due to rotational and translational non-crystallographic symmetry in human liver fructose-1,6-bisphosphatase | | Descriptor: | CHLORIDE ION, Fructose-1,6-bisphosphatase 1, SULFATE ION, ... | | Authors: | Ruf, A, Tetaz, T, Schott, B, Joseph, C, Rudolph, M.G. | | Deposit date: | 2016-06-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quadruple space-group ambiguity owing to rotational and translational noncrystallographic symmetry in human liver fructose-1,6-bisphosphatase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6ZSX
 
 | |
6ZS7
 
 | | Crystal structure of delta466-491 cystathionine beta-synthase from Toxoplasma gondii with L-cysteine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine beta-synthase | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the unique conformation of cystathionine beta-synthase from Toxoplasma gondii .
Comput Struct Biotechnol J, 19, 2021
|
|
5LYU
 
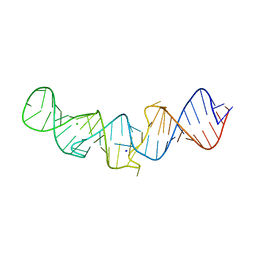 | | The native crystal structure of 7SK 5'-hairpin | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
6SR3
 
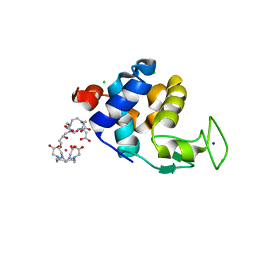 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 62 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6ZTC
 
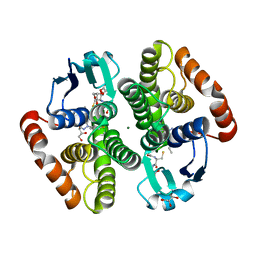 | | CRYSTAL STRUCTURE OF PROSTAGLANDIN D2 SYNTHASE IN COMPLEX WITH FRAGMENT 1A AT 1.84A RESOLUTION. | | Descriptor: | 1-[1-(3-fluorophenyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]propan-1-one, GLUTATHIONE, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
6ZU0
 
 | |
6SRA
 
 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with naringenin | | Descriptor: | GLUTATHIONE, R-naringenin, glutathione transferase | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SRN
 
 | |
5LEZ
 
 | | Human 20S proteasome complex with Oprozomib in Mg-Acetate at 2.2 Angstrom | | Descriptor: | ACETATE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
6SMR
 
 | | A. thaliana serine hydroxymethyltransferase isoform 4 (AtSHMT4) in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, METHOTREXATE, ... | | Authors: | Ruszkowski, M, Sekula, B, Dauter, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of methotrexate and pemetrexed action on serine hydroxymethyltransferases revealed using plant models.
Sci Rep, 9, 2019
|
|
6ZUE
 
 | |
6ST4
 
 | |
5LSO
 
 | |
6ZSK
 
 | |
6STC
 
 | |
6ZSW
 
 | |
1MC9
 
 | | STREPROMYCES LIVIDANS XYLAN BINDING DOMAIN CBM13 IN COMPLEX WITH XYLOPENTAOSE | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, SULFATE ION, ... | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2002-08-06 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
