1TUD
 
 | |
1TUC
 
 | |
1FKW
 
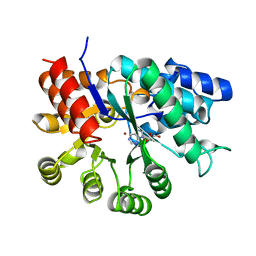 | | MURINE ADENOSINE DEAMINASE (D295E) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
1FKX
 
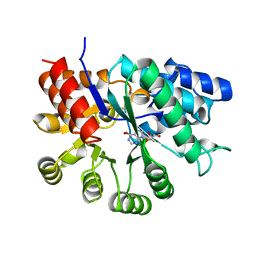 | | MURINE ADENOSINE DEAMINASE (D296A) | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
1ISO
 
 | |
3SSI
 
 | |
1GNH
 
 | | HUMAN C-REACTIVE PROTEIN | | Descriptor: | C-REACTIVE PROTEIN, CALCIUM ION | | Authors: | Shrive, A.K, Cheetham, G.M.T, Holden, D, Myles, D.A, Turnell, W.G, Volanakis, J.E, Pepys, M.B, Bloomer, A.C, Greenhough, T.J. | | Deposit date: | 1996-03-01 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three dimensional structure of human C-reactive protein.
Nat.Struct.Biol., 3, 1996
|
|
251D
 
 | |
1ODQ
 
 | |
1ODP
 
 | |
1ODR
 
 | |
4UPJ
 
 | |
1IPW
 
 | |
1VCQ
 
 | |
3UPJ
 
 | |
1VCP
 
 | |
1BHG
 
 | | HUMAN BETA-GLUCURONIDASE AT 2.6 A RESOLUTION | | Descriptor: | BETA-GLUCURONIDASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jain, S, Drendel, W.B. | | Deposit date: | 1996-03-04 | | Release date: | 1997-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs.
Nat.Struct.Biol., 3, 1996
|
|
1UPJ
 
 | |
2UPJ
 
 | |
1EHC
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN CHEY | | Descriptor: | CHEY, SULFATE ION | | Authors: | Jiang, M, Bourret, R, Simon, M, Volz, K. | | Deposit date: | 1996-03-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY.
J.Biol.Chem., 272, 1997
|
|
1PPR
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE | | Descriptor: | CHLOROPHYLL A, DIGALACTOSYL DIACYL GLYCEROL (DGDG), PERIDININ, ... | | Authors: | Hofmann, E, Welte, W, Diederichs, K. | | Deposit date: | 1996-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae.
Science, 272, 1996
|
|
1CED
 
 | | THE STRUCTURE OF CYTOCHROME C6 FROM MONORAPHIDIUM BRAUNII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Quacquarini, G, Walter, O, Diaz, A, Hervas, M, De La Rosa, M.A. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of cytochrome c6 from the green alga Monoraphidium braunii
J.Biol.Inorg.Chem., 1, 1996
|
|
1GAI
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
1GAH
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
252D
 
 | |
