4QRQ
 
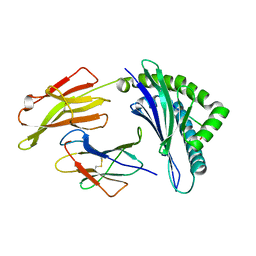 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
5T7G
 
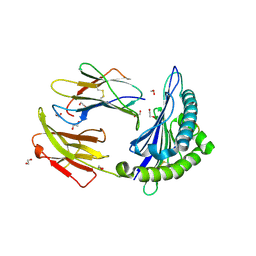 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PT9) of HIV gp120 MN Isolate (IGPGRAFYT) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-09-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
2OJT
 
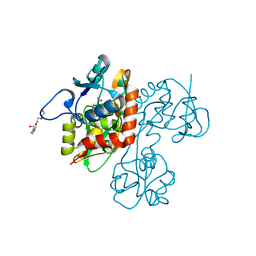 | | Structure and mechanism of kainate receptor modulation by anions | | Descriptor: | (S)-1-(2-AMINO-2-CARBOXYETHYL)-3(2-CARBOXYTHIOPHENE-3-YL-METHYL)-5-METHYLPYRIMIDINE-2,4-DIONE, BROMIDE ION, Glutamate receptor, ... | | Authors: | Mayer, M.L. | | Deposit date: | 2007-01-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of kainate receptor modulation by anions.
Neuron, 53, 2007
|
|
2QEJ
 
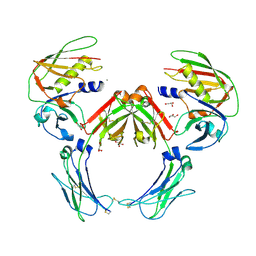 | | Crystal structure of a Staphylococcus aureus protein (SSL7) in complex with Fc of human IgA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ramsland, P.A, Willoughby, N, Trist, H.M, Farrugia, W, Hogarth, P.M, Fraser, J.D, Wines, B.D. | | Deposit date: | 2007-06-26 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2KSJ
 
 | | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Marassi, F, Black, D, Opella, S.J. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly.
Biophys.J., 99, 2010
|
|
4TVP
 
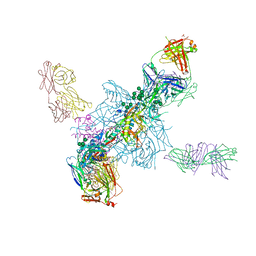 | | Crystal Structure of the HIV-1 BG505 SOSIP.664 Env Trimer Ectodomain, Comprising Atomic-Level Definition of Pre-Fusion gp120 and gp41, in Complex with Human Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Pancera, M, Zhou, T, Kwong, P.D. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and immune recognition of trimeric pre-fusion HIV-1 Env.
Nature, 514, 2014
|
|
1ES0
 
 | |
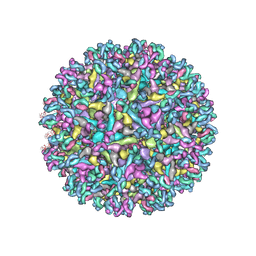
6XOB
 
 | |
6XLZ
 
 | | Structure of NHP D11A.F2 Fab in complex with 16055 V2b peptide | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Envelope glycoprotein gp160, NHP_D11A.F2_Fab_Heavy_chain, ... | | Authors: | Aljedani, S, Rodarte, J, Liban, T, Pancera, M. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
6XSN
 
 | |
6Y1L
 
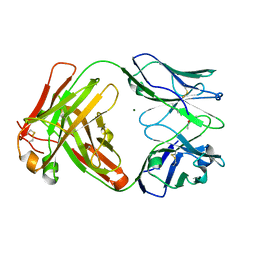 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y49
 
 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1N
 
 | | Crystal structure of the phosphonate-modified A.5 antibody FAB fragment | | Descriptor: | 8-METHYL-8-AZABICYCLO[3.2.1]OCTAN-3-YL PHENYLPHOSPHONATE, FAB A.5 Heavy chain, FAB A.5 Light Chain | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1K
 
 | | Crystal structure of the unmodified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | FAB A.17 L47R mutant Heavy Chain, FAB A.17 L47R mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y5Q
 
 | | human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6YM0
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | Descriptor: | Spike glycoprotein, heavy chain, light chain | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Y50
 
 | | 5'domain of human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Y1M
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47K mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light CHAIN | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLA
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z06
 
 | | Crystal structure of Puumala virus Gc in complex with Fab 4G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, Fab 4G2 Heavy chain, ... | | Authors: | Rissanen, I.R, Stass, R, Krumm, S.A, Seow, J, Hulswit, R.J.G, Paesen, G.C, Hepojoki, J, Vapalahti, O, Lundkvist, A, Reynard, O, Volchkov, V, Doores, K.J, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein.
Elife, 9, 2020
|
|
7PHQ
 
 | | Structure of homo-dimeric Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMT-Nb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PIJ
 
 | | Structure of Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMTNb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PHP
 
 | | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, Multidrug resistance protein NorM, NabFab HC, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Niederer, M, Pardon, E, Steyaert, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
