9FQI
 
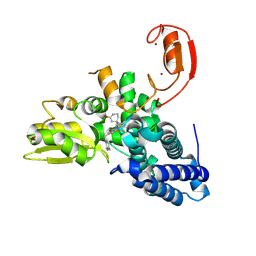 | | E3 ligase Cbl-b in complex with a lactam scaffold inhibitor (compound 7) | | Descriptor: | 8-[3-[(4~{R})-4-methyl-2-oxidanylidene-piperidin-4-yl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQJ
 
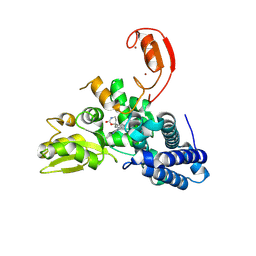 | | E3 ligase Cbl-b in complex with a carbamate scaffold inhibitor (compound 12) | | Descriptor: | 2-cyclopropyl-6-methyl-~{N}-[3-[(6~{S})-6-methyl-2-oxidanylidene-1,3-oxazinan-6-yl]phenyl]pyrimidine-4-carboxamide, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.563 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQH
 
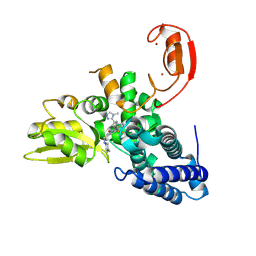 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | Descriptor: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FWX
 
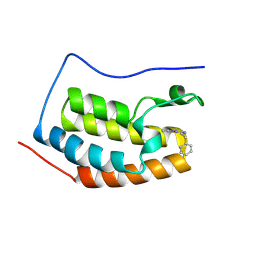 | | Crystal structure of BRD4 BD1 with MEN1404BS. | | Descriptor: | 3-methyl-~{N}-(2-phenylethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D. | | Deposit date: | 2024-07-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
9FXP
 
 | | Crystal structure of BRD4 BD1 with DI00626383. | | Descriptor: | 4-methoxy-1,2-benzoxazol-3-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Reinert, D. | | Deposit date: | 2024-07-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
4G34
 
 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | Descriptor: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4G31
 
 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
5DB2
 
 | | Menin in complex with MI-389 | | Descriptor: | 2-{2-cyano-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indol-1-yl}aceta mide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DDE
 
 | | Menin in complex with MI-859 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2,2-difluoroethyl)-4-[4-(5,5-dimethyl-4,5-dihydro-1,3-thiazol-2-yl)piperazin-1-yl]thieno[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Rational Design of Orthogonal Multipolar Interactions with Fluorine in Protein-Ligand Complexes.
J.Med.Chem., 58, 2015
|
|
5DB1
 
 | | Menin in complex with MI-336 | | Descriptor: | 6-methoxy-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
9GIL
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound 12 | | Descriptor: | (7~{R},11~{R},19~{E})-11-[(4-chlorophenyl)methyl]-13-oxa-3,10,23-triazatricyclo[19.3.1.0^{3,7}]pentacosa-1(24),19,21(25),22-tetraene-2,9,12-trione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Schmitt, A, Preuss, F, Prasad, A, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
9GIJ
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound 5 | | Descriptor: | (2~{R})-3-(4-chlorophenyl)-2-[2-[(2~{R})-1-isoquinolin-4-ylcarbonylpyrrolidin-2-yl]ethanoyl-methyl-amino]-~{N}-methyl-propanamide, 3C-like proteinase nsp5 | | Authors: | Prasad, A, Schmitt, A, Preuss, F, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
6CI6
 
 | | Crystal structure of equine serum albumin in complex with nabumetone | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Serum albumin, ... | | Authors: | Venkataramany, B.S, Czub, M.P, Shabalin, I.G, Handing, K.B, Steen, E.H, Cooper, D.R, Joachimiak, A, Satchell, K.J.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 2020
|
|
6QJ5
 
 | | X-ray structure of PPARgamma LBD with the ligand NV1380 | | Descriptor: | (2~{S})-3-methyl-2-[(4-octoxyphenyl)carbonylamino]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D. | | Deposit date: | 2019-01-22 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor gamma Binding Properties and Biological Activities.
J.Med.Chem., 63, 2020
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 67, 2024
|
|
5DKR
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
7B9Y
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 64a | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,24(28),25-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7BA0
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 63 | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,28-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.2.2.113,17.04,9]nonacosa-1(26),13(29),14,16,24,27-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B9Z
 
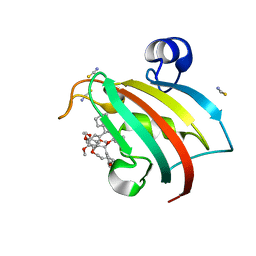 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 35-(E) | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,20,24(28),25-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5, isothiocyanate | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6JZ0
 
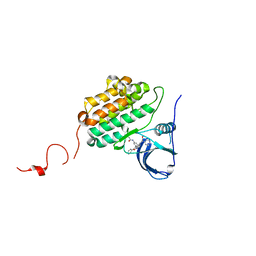 | | Crystal structure of EGFR kinase domain in complex with compound 78 | | Descriptor: | E-4-(dimethylamino)-N-[3-[4-[[(1S)-2-oxidanyl-1-phenyl-ethyl]amino]-6-phenyl-furo[2,3-d]pyrimidin-5-yl]phenyl]but-2-enamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a Furanopyrimidine-Based Epidermal Growth Factor Receptor Inhibitor (DBPR112) as a Clinical Candidate for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 62, 2019
|
|
9FBX
 
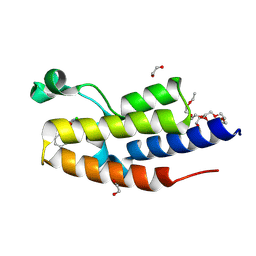 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one, ... | | Authors: | Chung, C. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structure- and Property-Based Optimization of Efficient Pan-Bromodomain and Extra Terminal Inhibitors to Identify Oral and Intravenous Candidate I-BET787.
J.Med.Chem., 67, 2024
|
|
9CSI
 
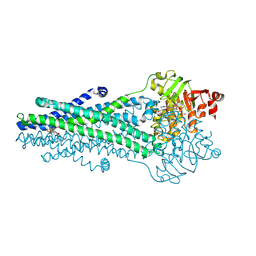 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9FO2
 
 | | Coxsackievirus A9 bound with compound 15 (CL278) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
9FKZ
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(4-chlorophenyl)methyl]prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9FKY
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(1~{S})-1-[3-cyano-4-(trifluoromethyl)phenyl]ethyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
