1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
1N3O
 
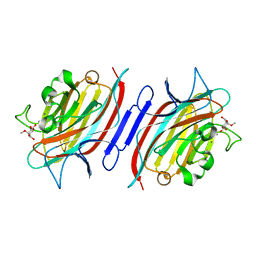 | | Pterocarcpus angolensis lectin in complex with alpha-methyl glucose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin PAL, ... | | Authors: | Loris, R, Imberty, A, Beeckmans, S, Van Driessche, E, Read, J.S, Bouckaert, J, De Greve, H, Buts, L, Wyns, L. | | Deposit date: | 2002-10-29 | | Release date: | 2002-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Pterocarpus angolensis lectin in complex with glucose, sucrose, and turanose
J.BIOL.CHEM., 278, 2003
|
|
1X4E
 
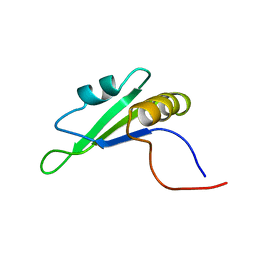 | | Solution structure of RRM domain in RNA binding motif, single-stranded interacting protein 2 | | Descriptor: | RNA binding motif, single-stranded interacting protein 2 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in RNA binding motif, single-stranded interacting protein 2
To be Published
|
|
1WE8
 
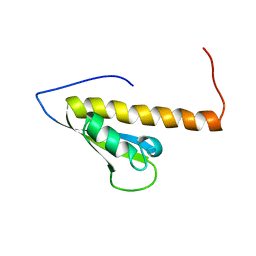 | | Solution structure of KH domain in protein BAB28342 | | Descriptor: | Tudor and KH domain containing protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in protein BAB28342
To be Published
|
|
1ZVT
 
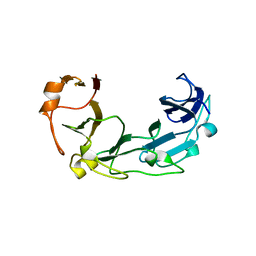 | |
1N37
 
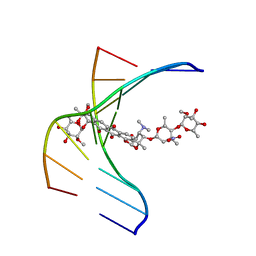 | |
212D
 
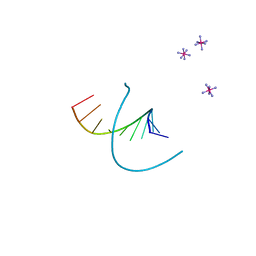 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
1ZRY
 
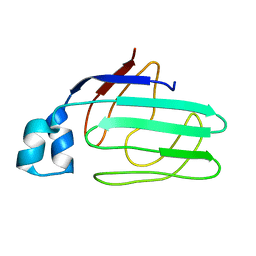 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
1WPK
 
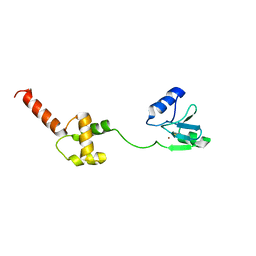 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1X4M
 
 | | Solution structure of KH domain in Far upstream element binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in Far upstream element binding protein 1
To be Published
|
|
1NIQ
 
 | |
1X1T
 
 | | Crystal Structure of D-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi Complexed with NAD+ | | Descriptor: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | Deposit date: | 2005-04-13 | | Release date: | 2006-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
1WH6
 
 | | Solution structure of the second CUT domain of human Homeobox protein Cux-2 | | Descriptor: | Homeobox protein Cux-2 | | Authors: | Nameki, N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second CUT domain of human Homeobox protein Cux-2
To be Published
|
|
2BIT
 
 | | Crystal structure of human cyclophilin D at 1.7 A resolution | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WHN
 
 | | Solution structure of the dsRBD from hypothetical protein BAB26260 | | Descriptor: | hypothetical protein RIKEN cDNA 2310016K04 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dsRBD from hypothetical protein BAB26260
To be Published
|
|
1WK0
 
 | | Solution structure of Fibronectin type III domain derived from human KIAA0970 protein | | Descriptor: | KIAA0970 protein | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Hayashi, F, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Fibronectin type III domain derived from human KIAA0970 protein
To be Published
|
|
1N8U
 
 | | Chemosensory Protein in Complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N8Y
 
 | | Crystal structure of the extracellular region of rat HER2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, protooncoprotein | | Authors: | Cho, H.-S, Mason, K, Ramyar, K.X, Stanley, A.M, Gabelli, S.B, Denney Jr, D.W, Leahy, D.J. | | Deposit date: | 2002-11-21 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Extracellular Region of HER2 Alone and in complex with the Herceptin Fab
Nature, 421, 2003
|
|
1N96
 
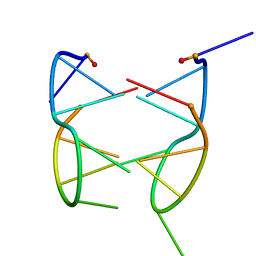 | | DIMERIC SOLUTION STRUCTURE OF THE CYCLIC OCTAMER CD(CGCTCATT) | | Descriptor: | CYCLIC OLIGONUCLEOTIDE D(CGCTCATT) | | Authors: | Escaja, N, Gelpi, J.L, Orozco, M, Rico, M, Pedroso, E, Gonzalez, C. | | Deposit date: | 2002-11-22 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad.
J.Am.Chem.Soc., 125, 2003
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
1WHX
 
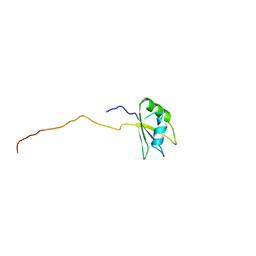 | | Solution structure of the second RNA binding domain from hypothetical protein BAB23448 | | Descriptor: | hypothetical protein RIKEN CDNA 1200009A02 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA binding domain from hypothetical protein BAB23448
To be Published
|
|
1NIJ
 
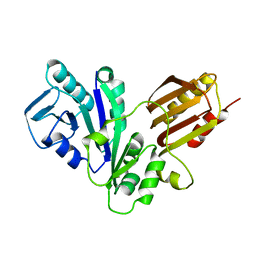 | | YJIA PROTEIN | | Descriptor: | Hypothetical protein yjiA | | Authors: | Khil, P.P, Obmolova, G, Teplyakov, A, Howard, A.J, Gilliland, G.L, Camerini-Otero, R.D, Structure 2 Function Project (S2F) | | Deposit date: | 2002-12-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.
Proteins, 54, 2004
|
|
1WV4
 
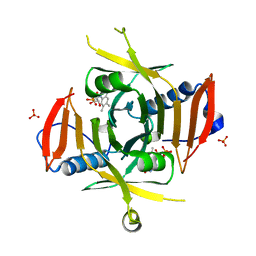 | | X-ray Structure of Escherichia coli pyridoxine 5'-phosphate oxidase in tetragonal crystal form | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, Pyridoxamine 5'-phosphate oxidase | | Authors: | Safo, M.K, Musayev, F.N, Schirch, V. | | Deposit date: | 2004-12-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli pyridoxine 5'-phosphate oxidase in a tetragonal crystal form: insights into the mechanistic pathway of the enzyme.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BLS
 
 | |
2BWO
 
 | | 5-Aminolevulinate Synthase from Rhodobacter capsulatus in complex with succinyl-CoA | | Descriptor: | 5-AMINOLEVULINATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SUCCINYL-COENZYME A | | Authors: | Astner, I, Schulze, J.O, van den Heuvel, J.J, Jahn, D, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-27 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 5-Aminolevulinate Synthase, the First Enzyme of Heme Biosynthesis, and its Link to Xlsa in Humans.
Embo J., 24, 2005
|
|
