2I45
 
 | |
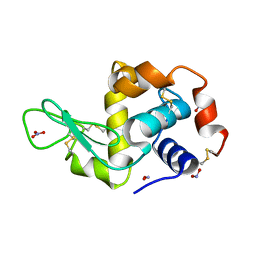
2YBI
 
 | |
2XA8
 
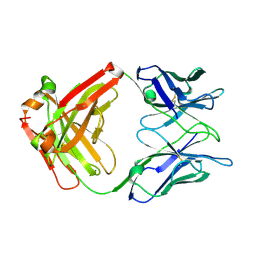 | | Crystal structure of the Fab domain of omalizumab at 2.41A | | Descriptor: | OMALIZUMAB HEAVY CHAIN, OMALIZUMAB LIGHT CHAIN | | Authors: | Huang, C.H, Hung, F.H.A, Lim, C, Chang, T.W, Ma, C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Physical Basis for Anti-IgE Therapy.
Sci Rep, 5, 2015
|
|
2WOJ
 
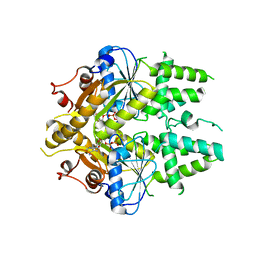 | | ADP-AlF4 complex of S. cerevisiae GET3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, MAGNESIUM ION, ... | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
2WJE
 
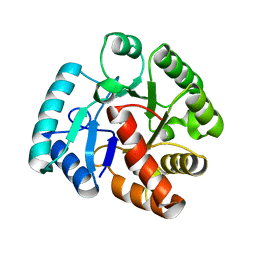 | |
2WJF
 
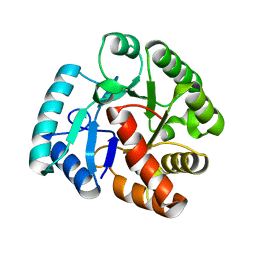 | | Crystal structure of the tyrosine phosphatase Cps4B from Steptococcus pneumoniae TIGR4 in complex with phosphate. | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, TYROSINE-PROTEIN PHOSPHATASE CPSB | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2009-05-25 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structures of Wzb of Escherichia Coli and Cpsb of Streptococcus Pneumoniae, Representatives of Two Families of Tyrosine Phosphatases that Regulate Capsule Assembly.
J.Mol.Biol., 392, 2009
|
|
2I24
 
 | |
2XA7
 
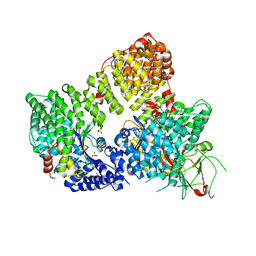 | | AP2 clathrin adaptor core in active complex with cargo peptides | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, ALPHA 2 SUBUNIT, AP-2 COMPLEX SUBUNIT BETA, ... | | Authors: | Jackson, L.P, Kelly, B.T, McCoy, A.J, Evans, P.R, Owen, D.J. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Large Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the Ap2 Clathrin Adaptor Complex
Cell(Cambridge,Mass.), 141, 2010
|
|
2WX3
 
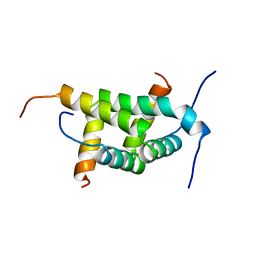 | |
2X8K
 
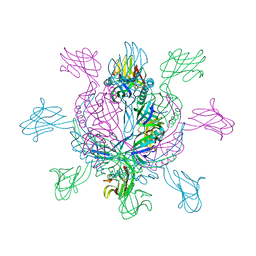 | | Crystal Structure of SPP1 Dit (gp 19.1) Protein, a Paradigm of Hub Adsorption Apparatus in Gram-positive Infecting Phages. | | Descriptor: | HYPOTHETICAL PROTEIN 19.1 | | Authors: | Veesler, D, Robin, G, Lichiere, J, Auzat, I, Tavares, P, Bron, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Bacteriophage Spp1 Distal Tail Protein (Gp 19.1): A Baseplate Hub Paradigm in Gram+ Infecting Phages.
J.Biol.Chem., 285, 2010
|
|
2IBG
 
 | |
2I6X
 
 | | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. (CASP Target)
To be Published
|
|
2HLV
 
 | | Bovine Odorant Binding Protein deswapped triple mutant | | Descriptor: | 3,6-BIS(METHYLENE)DECANOIC ACID, GLYCEROL, Odorant-binding protein | | Authors: | Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2006-07-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deswapping bovine odorant binding protein.
Biochim.Biophys.Acta, 1784, 2008
|
|
2WJ6
 
 | |
2HUK
 
 | |
2HWG
 
 | | Structure of phosphorylated Enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system | | Descriptor: | MAGNESIUM ION, OXALATE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Lim, K, Teplyakov, A, Herzberg, O. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2XFV
 
 | | Structure of the amino-terminal domain from the cell-cycle regulator Swi6 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Smerdon, S.J, Goldstone, D.C, Taylor, I.A. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Amino-Terminal Domain from the Cell-Cycle Regulator Swi6S
Proteins, 78, 2010
|
|
2XGX
 
 | | Crystal structure of transcription factor NtcA from Synechococcus elongatus (mercury derivative) | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLOBAL NITROGEN REGULATOR, ... | | Authors: | Llacer, J.L, Castells, M.A, Rubio, V. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Regulation of Ntca-Dependent Transcription by Proteins Pipx and Pii.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2YH3
 
 | | The structure of BamB from E. coli | | Descriptor: | LIPOPROTEIN YFGL | | Authors: | Albrecht, R, Zeth, K. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
2Y7P
 
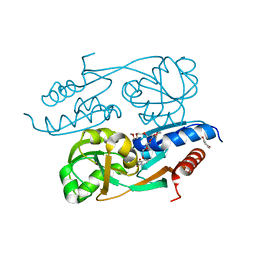 | | DntR Inducer Binding Domain in Complex with Salicylate. Trigonal crystal form | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-HYDROXYBENZOIC ACID, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Devesse, L, Smirnova, I, Lonneborg, R, Kapp, U, Brzezinski, P, Leonard, G.A, Dian, C. | | Deposit date: | 2011-02-01 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Dntr Inducer Binding Domains in Complex with Salicylate Offer Insights Into the Activation of Lysr-Type Transcriptional Regulators.
Mol.Microbiol., 81, 2011
|
|
2HYX
 
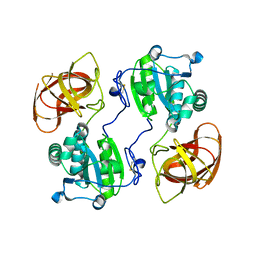 | |
2YMU
 
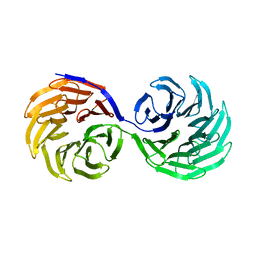 | |
2HU0
 
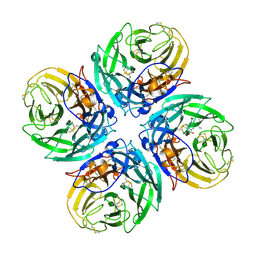 | | N1 neuraminidase in complex with oseltamivir 1 | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HTY
 
 | | N1 neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2YL2
 
 | | Crystal structure of human acetyl-CoA carboxylase 1, biotin carboxylase (BC) domain | | Descriptor: | ACETYL-COA CARBOXYLASE 1 | | Authors: | Muniz, J.R.C, Froese, D.S, Krysztofinska, E, Vollmar, M, Beltrami, A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Acetyl-Coa Carboxylase 1, Biotin Carboxylase (Bc) Domain
To be Published
|
|
