7SMH
 
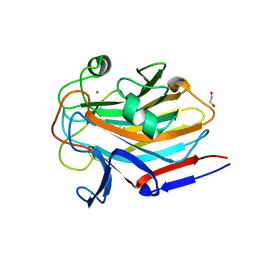 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
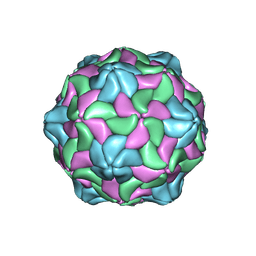
6P4L
 
 | |
7PYB
 
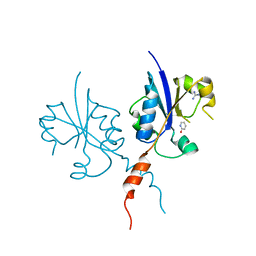 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-Hydroxypyridine | | Descriptor: | 1~{H}-pyridin-2-one, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-09 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
6P7J
 
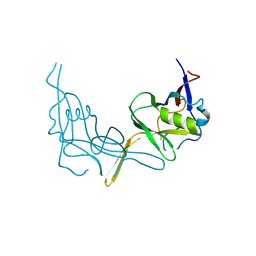 | |
7D7Z
 
 | |
6XCZ
 
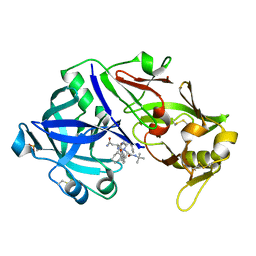 | | Porcine pepsin in complex with saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Pepsin A | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Porcine pepsin in complex with saquinavir
To Be Published
|
|
6P1B
 
 | | Transcription antitermination factor Q21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Q protein | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P88
 
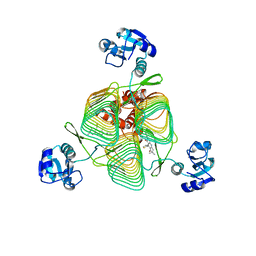 | | E.coli LpxD in complex with compound 6 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-{3-[(furan-2-carbonyl)amino]phenyl}-2,3-dihydro-1,4-benzodioxine-6-carboxamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
7Q2I
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Tetrahydrofurfurylamine | | Descriptor: | 1-[(2R)-oxolan-2-yl]methanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8XQU
 
 | | The Crystal Structure of ClpC1-NTD from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease ATP-binding subunit ClpC1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of ClpC1-NTD from Biortus.
To Be Published
|
|
1JZV
 
 | | Crystal structure of a bulged RNA from the SL2 stem-loop of the HIV-1 psi-RNA | | Descriptor: | 5'-R(*CP*AP*GP*UP*AP*CP*GP*(5IC)P*C)-3', 5'-R(*GP*GP*CP*GP*AP*CP*(5BU)P*G)-3', MAGNESIUM ION | | Authors: | Xiong, Y, Sudarsanakumar, C, Deng, J, Pan, B, Sundaralingam, M. | | Deposit date: | 2001-09-17 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Bulged RNA from the SL2 Stem-loop of the HIV-1 psi-RNA
To be Published
|
|
6ZXV
 
 | |
4Y9D
 
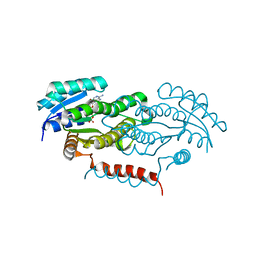 | | Crystal structure of LigD in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
6FRA
 
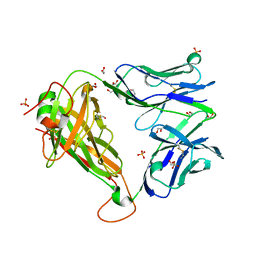 | |
7A7P
 
 | | rsGreen0.7-K206A partially in the green-off state | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, NITRATE ION | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8VA0
 
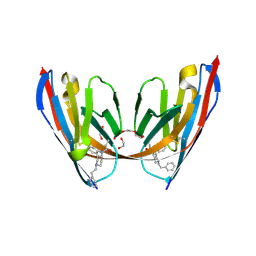 | | X-ray crystal structure of JGFN4 N76D complexed with fentanyl in dimer form | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9X
 
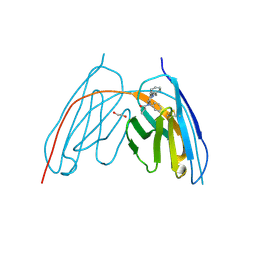 | | X-ray crystal structure of JGFN4 complex with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8C6U
 
 | | PBP AccA-G145YG440Q mutant from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8Q29
 
 | |
7TDB
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, manganese ions | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
9EQQ
 
 | |
7TDC
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, calcium ions | | Descriptor: | CALCIUM ION, MAGNESIUM ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
8RQG
 
 | |
8YK7
 
 | | Structure of Rib domain from surface adhesin of Limosilactobacillus reuteri | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the Rib domain of the cell-wall-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
4ZC8
 
 | | Structure of Acetobacter aceti PurE-S57T | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N5-carboxyaminoimidazole ribonucleotide mutase, ... | | Authors: | Kappock, T.J, Sullivan, K.L. | | Deposit date: | 2015-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Mutation of the conserved serine in two PurE classes
To Be Published
|
|
