6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | Authors: | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6N64
 
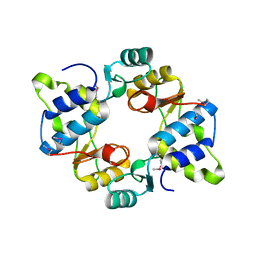 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
7MR5
 
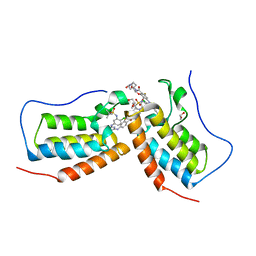 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-052 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-{oxybis[(ethane-2,1-diyl)oxy(1-oxoethane-2,1-diyl)piperidine-1,4-diyl]}bis{4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluorobenzamide} | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
8EKR
 
 | | Apo rat TRPV2 in nanodiscs, state 3 | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional and structural insights into activation of TRPV2 by weak acids.
Embo J., 43, 2024
|
|
4WA4
 
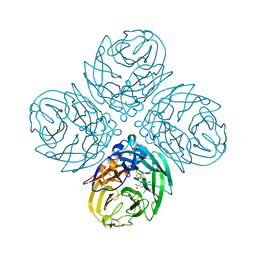 | | The crystal structure of neuraminidase from a H3N8 influenza virus isolated from New England harbor seals in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
8EKQ
 
 | | Apo rat TRPV2 in nanodiscs, state 2 | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Functional and structural insights into activation of TRPV2 by weak acids.
Embo J., 43, 2024
|
|
6W4V
 
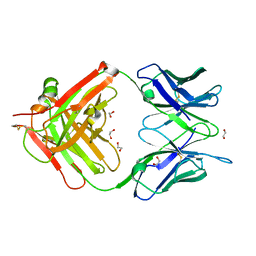 | | Structure of anti-ferroportin Fab45D8 | | Descriptor: | 1,2-ETHANEDIOL, Fab45D8 Heavy Chain, Fab45D8 Light Chain | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
4W5M
 
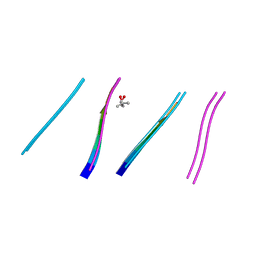 | | Prp peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
7O04
 
 | |
4W5Y
 
 | | Crystal structure of Prp pepttide | | Descriptor: | Prp peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-19 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.122 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
8HNQ
 
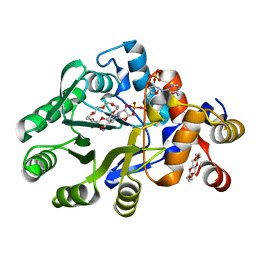 | | The structure of a alcohol dehydrogenase AKR13B2 with NADP | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chen, M, Yang, H, Lu, F. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of AKR13B2
To Be Published
|
|
7MME
 
 | |
6VXI
 
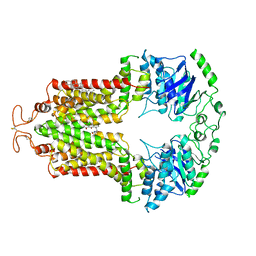 | | Structure of ABCG2 bound to mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
4YCD
 
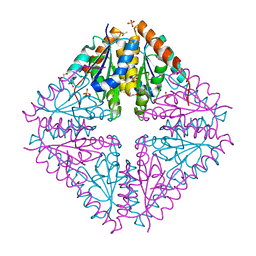 | |
8EI8
 
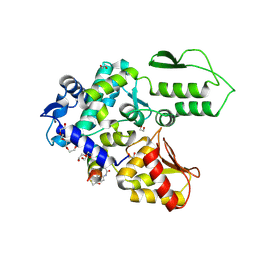 | | Crystal structure of the WWP2 HECT domain in complex with H308, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, H308, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8YGV
 
 | | F1 domain of Non-catalytic site depleted and epsilon C-terminal domain deleted FoF1-ATPase from Bacillus PS3,nucleotide depleted condition | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Kobayashi, R, Nakano, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
7OA0
 
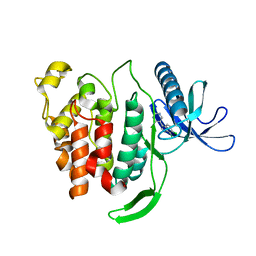 | | Crystal structure of di-phosphorylated human CLK1 in complex with 5-(6,7-dichloro-1H-indol-3-yl)pyrimidin-4-amine | | Descriptor: | 5-[6,7-bis(chloranyl)-1-methyl-indol-3-yl]pyrimidin-4-amine, Dual specificity protein kinase CLK1 | | Authors: | Livnah, O, Domovich, Y, Bracher, F, Aiger, C. | | Deposit date: | 2021-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Development of novel CLK1 inhibitors as potential drugs for treatment of Chikungunya virus infections
To Be Published
|
|
6Y1G
 
 | | Photoconverted HcRed in its optoacoustic state | | Descriptor: | 1,2-ETHANEDIOL, GFP-like non-fluorescent chromoprotein | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-02-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Challenging a Preconception: Optoacoustic Spectrum Differs from the Optical Absorption Spectrum of Proteins and Dyes for Molecular Imaging.
Anal.Chem., 92, 2020
|
|
6NSJ
 
 | | CryoEM structure of Helicobacter pylori urea channel in closed state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
8YH8
 
 | | F1 domain of Non-catalytic site depleted and epsilon C-terminal domain deleted FoF1-ATPase from Bacillus PS3,under ATP saturated condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Kobayashi, R, Nakano, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2024-02-27 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | ADP-inhibited structure of non-catalytic site-depleted F o F 1 -ATPase from thermophilic Bacillus sp. PS-3.
Biochim Biophys Acta Bioenerg, 1866, 2025
|
|
6NBO
 
 | |
8F97
 
 | | Compound 5 bound to procaspase-6 | | Descriptor: | 1,2-ETHANEDIOL, 2-({[(2M)-[2,3'-bipyridin]-2'-yl]amino}methyl)-5-fluorophenol, CHLORIDE ION, ... | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8F9D
 
 | | Compound 21 bound to procaspase-6 | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoro-2-({[(3M)-3-(1,2,4-oxadiazol-3-yl)pyridin-2-yl]amino}methyl)phenol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
6YPH
 
 | | Crystal Structure of CK2alpha with Compound 2 bound | | Descriptor: | 4-[(4-naphthalen-2-yl-1,3-thiazol-2-yl)amino]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
7OKV
 
 | | Crystal structure of soluble EPCR after exposure to the nonionic surfactant Polysorbate 20 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Erausquin, E, Dichiara, M.G, Lopez-Sagaseta, J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of a broad lipid repertoire associated to the endothelial cell protein C receptor (EPCR).
Sci Rep, 12, 2022
|
|
