7XDH
 
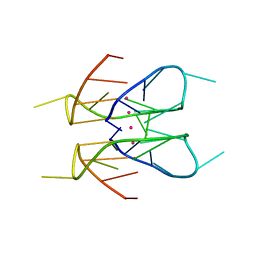 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
8EJP
 
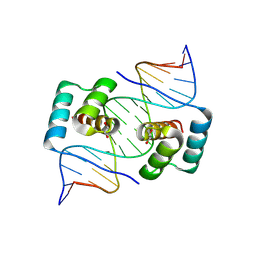 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
7MOW
 
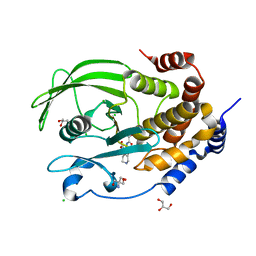 | | PTP1B F225I in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MOU
 
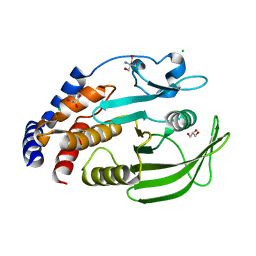 | | PTP1B F225Y-R199N-L195R | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
8RD3
 
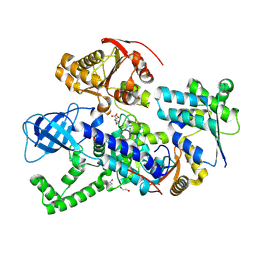 | |
7SUA
 
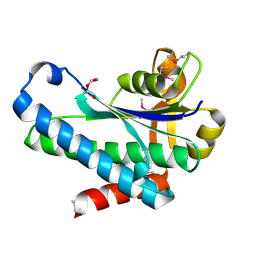 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
7MRC
 
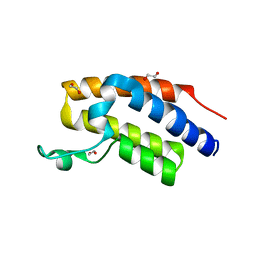 | |
8ER5
 
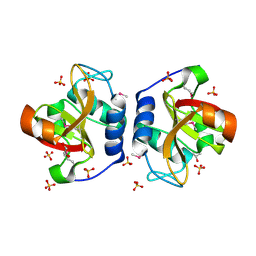 | | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NlpC/P60 domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448.
To Be Published
|
|
8HOE
 
 | |
7O9Y
 
 | |
6NFH
 
 | | BTK in complex with inhibitor 8-(2,3-dihydro-1H-inden-5-yl)-2-({4-[(2S)-3-(dimethylamino)-2-hydroxypropoxy]phenyl}amino)-5,8-dihydropteridine-6,7-dione | | Descriptor: | 1,2-ETHANEDIOL, 8-(2,3-dihydro-1H-inden-5-yl)-2-({4-[(2S)-3-(dimethylamino)-2-hydroxypropoxy]phenyl}amino)-5,8-dihydropteridine-6,7-dione, Tyrosine-protein kinase BTK | | Authors: | Mirzadegan, T, Damm-Ganamet, K.L. | | Deposit date: | 2018-12-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Accelerating Lead Identification by High Throughput Virtual Screening: Prospective Case Studies from the Pharmaceutical Industry.
J.Chem.Inf.Model., 59, 2019
|
|
6W1X
 
 | | Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
6NKG
 
 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
6WAX
 
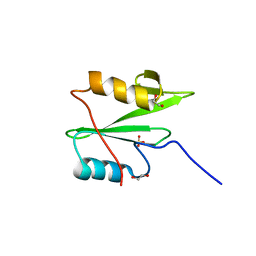 | | C-terminal SH2 domain of p120RasGAP | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein 1, SULFATE ION | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
7MXQ
 
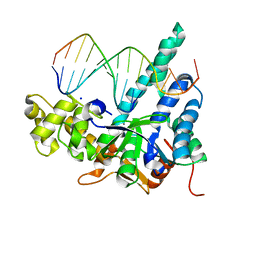 | |
6W70
 
 | | Crystal Structure of apixaban-bound ABLE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, ACETATE ION, De novo designed ABLE, ... | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6NE9
 
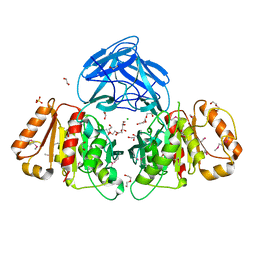 | | Bacteroides intestinalis acetyl xylan esterase (BACINT_01039) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Pereira, G.V, Cann, I. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Degradation of complex arabinoxylans by human colonic Bacteroidetes
Nat Commun, 2021
|
|
7NBQ
 
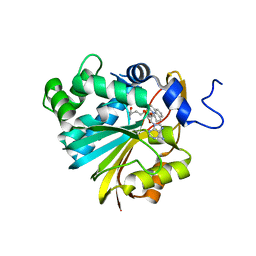 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with the tricyclic inhibitor (4) | | Descriptor: | 2-methyl-1,2,6,7-tetrahydro-3H,5H-pyrido[3,2,1-ij]quinazolin-3-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2021-01-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Molecules, 26, 2021
|
|
7OAS
 
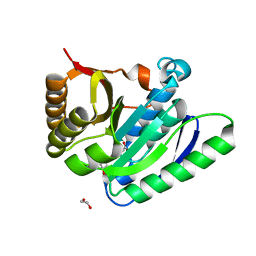 | | Structural basis for targeted p97 remodeling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XHD
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A•G•G•G•G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7NF5
 
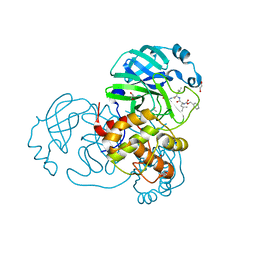 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
4WHK
 
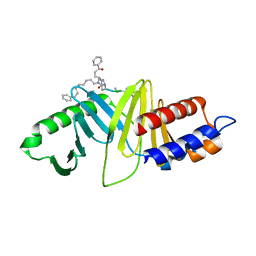 | | A New Class of Peptidomimetics Targeting the Polo-box Domain of Polo-like kinase 1 | | Descriptor: | C6H5(CH2)8-DERIVATIZED PEPTIDE INHIBITOR, Serine/threonine-protein kinase PLK1 | | Authors: | Bang, J.K, Han, Y.H, Ahn, M.J, Lee, K.S. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
8Z9A
 
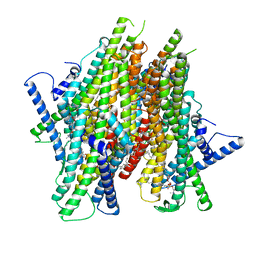 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
9CJM
 
 | | Red fluorescent protein mRuby3 | | Descriptor: | 1,2-ETHANEDIOL, Red fluorescent protein mRuby3 | | Authors: | Huang, M, Ng, H.L, Geisbrecht, B.V, Duan, H, Deng, M, Chu, J. | | Deposit date: | 2024-07-07 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal model of the red fluorescent protein mRuby3
To Be Published
|
|
8ZBO
 
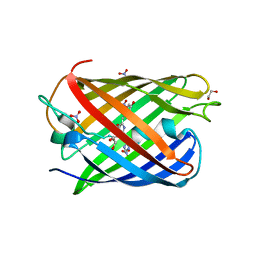 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
