8H7X
 
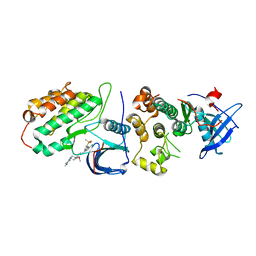 | | Crystal structure of EGFR T790M/C797S mutant in complex with brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
NPJ Precis Oncol, 8, 2024
|
|
6XVN
 
 | |
6XVM
 
 | |
6XVO
 
 | |
6XX4
 
 | |
6XX5
 
 | |
6XX2
 
 | |
6XX3
 
 | |
6WQW
 
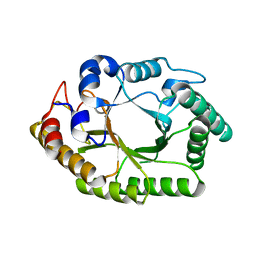 | | Thermobacillus composti GH10 xylanase | | Descriptor: | Beta-xylanase, beta-D-xylopyranose | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Transformation of xylan into value-added biocommodities using Thermobacillus composti GH10 xylanase.
Carbohydr Polym, 247, 2020
|
|
1IOP
 
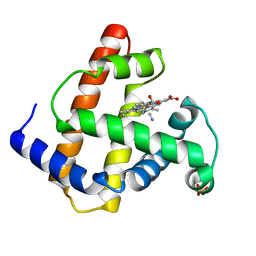 | | INCORPORATION OF A HEMIN WITH THE SHORTEST ACID SIDE-CHAINS INTO MYOGLOBIN | | Descriptor: | 6,7-DICARBOXYL-1,2,3,4,5,8-HEXAMETHYLHEMIN, CYANIDE ION, MYOGLOBIN, ... | | Authors: | Igarashi, N, Neya, S, Funasaki, N, Tanaka, N. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of 6,7-dicarboxyheme-substituted myoglobin
Biochemistry, 37, 1998
|
|
8GDV
 
 | |
8AH7
 
 | |
7X15
 
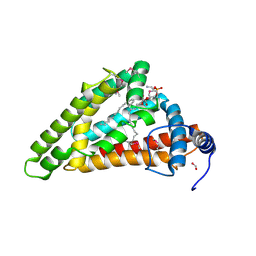 | | Crystal structure of MIGA2 LD targeting domain | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, FORMIC ACID, Mitoguardin 2 | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7X14
 
 | | Crystal structure of phospho-FFAT motif of MIGA2 bound to VAPB | | Descriptor: | MIGA2 phospho FFAT motif, SULFATE ION, Vesicle-associated membrane protein-associated protein B | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
5LTV
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
8AH5
 
 | |
5LTX
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
8AH6
 
 | |
8AH8
 
 | |
8AH4
 
 | |
6S7N
 
 | |
5MTW
 
 | |
6QDB
 
 | |
6QDA
 
 | | Leishmania major N-myristoyltransferase in complex with quinazoline inhibitor IMP-0000811 | | Descriptor: | 3-[methyl-[2-[methyl-(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDG
 
 | | Leishmania major N-myristoyltransferase in complex with quinazoline inhibitor IMP-0000169 | | Descriptor: | 3-[[6-bromanyl-2-[3-(dimethylamino)propyl-methyl-amino]quinazolin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
