6MMJ
 
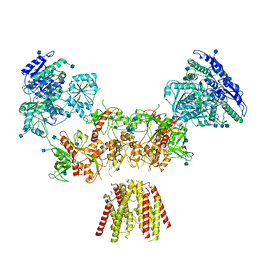 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Super-Splayed' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6GXL
 
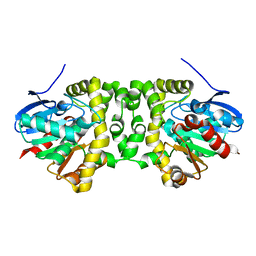 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: RADDAM2 | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
5V6U
 
 | |
6MMK
 
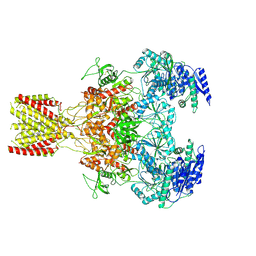 | | Diheteromeric NMDA receptor GluN1/GluN2A in the '1-Knuckle' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMH
 
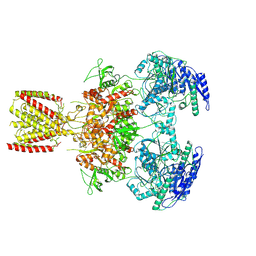 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Extended-2' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.21 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMW
 
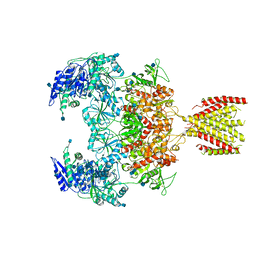 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MPI
 
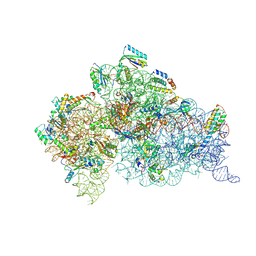 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-10-07 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
7XZ9
 
 | |
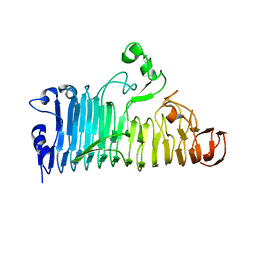
7XZD
 
 | |
7XZF
 
 | | Wild type of the N-terminal domain of fucoidan lyase FdlA | | Descriptor: | Fucoidan lyase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Li, M, Pan, X. | | Deposit date: | 2022-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analysis Reveals Catalytic Mechanism of Fucoidan Lyase from Flavobacterium sp. SA-0082.
Mar Drugs, 20, 2022
|
|
7XZ8
 
 | |
7K80
 
 | | KIR3DL1*001 in complex with HLA-A*24:02 presenting the RYPLTFGW peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ARG-TYR-PRO-LEU-THR-PHE-GLY-TRP, ... | | Authors: | MacLachlan, B.J, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Role of the HLA Class I alpha 2 Helix in Determining Ligand Hierarchy for the Killer Cell Ig-like Receptor 3DL1.
J Immunol., 206, 2021
|
|
7XZB
 
 | |
7XZA
 
 | |
7XZ7
 
 | |
7XZE
 
 | |
7XZC
 
 | |
6MM9
 
 | | Diheteromeric NMDA receptor GluN1/GluN2A in the '1-Knuckle' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 6.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-29 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMU
 
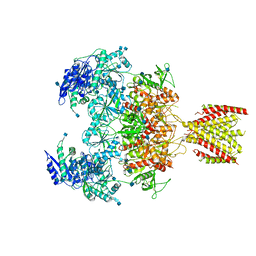 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '2-Knuckle-Asymmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
7YEP
 
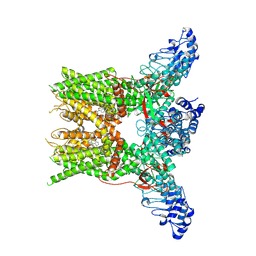 | | 2-APB bound state of mTRPV2 | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
6NUI
 
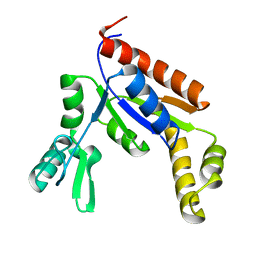 | | Human Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Sabo, T.M, Khan, N, Ban, D, Trigo-Mourino, P, Carneiro, M.G, Trent, J.O, Konrad, M, Lee, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional investigation of human guanylate kinase reveals allosteric networking and a crucial role for the enzyme in cancer.
J.Biol.Chem., 294, 2019
|
|
5VXC
 
 | |
5T4N
 
 | |
6O01
 
 | |
6NRL
 
 | |
