1DIS
 
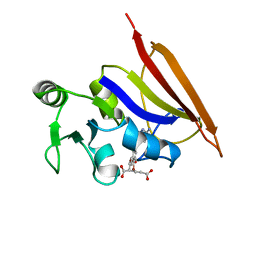 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
1DIT
 
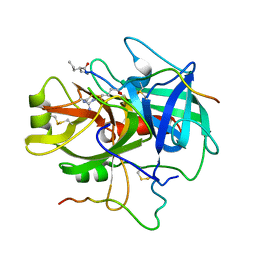 | | COMPLEX OF A DIVALENT INHIBITOR WITH THROMBIN | | Descriptor: | ALPHA-THROMBIN, PEPTIDE INHIBITOR CVS995 | | Authors: | Tulinsky, A, Krishnan, R. | | Deposit date: | 1995-07-20 | | Release date: | 1996-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, structure, and structure-activity relationships of divalent thrombin inhibitors containing an alpha-keto-amide transition-state mimetic.
Protein Sci., 5, 1996
|
|
1DIU
 
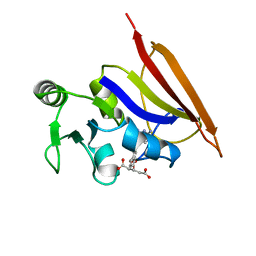 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
1DIV
 
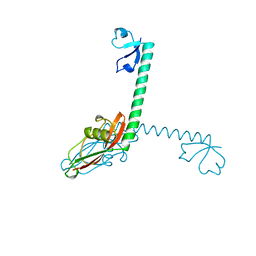 | | RIBOSOMAL PROTEIN L9 | | Descriptor: | RIBOSOMAL PROTEIN L9 | | Authors: | Hoffman, D.W, Cameron, C, Davies, C, Gerchman, S.E, Kycia, J.H, Porter, S, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein.
EMBO J., 13, 1994
|
|
1DIW
 
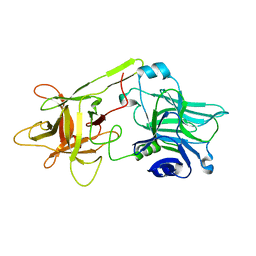 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH GALACTOSE | | Descriptor: | TETANUS TOXIN HC, beta-D-galactopyranose | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-11-30 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1DIX
 
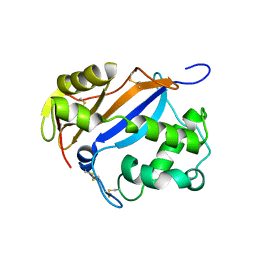 | | CRYSTAL STRUCTURE OF RNASE LE | | Descriptor: | EXTRACELLULAR RIBONUCLEASE LE | | Authors: | Tanaka, N, Nakamura, K.T. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a plant ribonuclease, RNase LE.
J.Mol.Biol., 298, 2000
|
|
1DIY
 
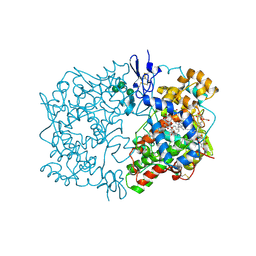 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND IN THE CYCLOOXYGENASE ACTIVE SITE OF PGHS-1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Malkowski, M.G, Ginell, S.L, Smith, W.L, Garavito, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The productive conformation of arachidonic acid bound to prostaglandin synthase.
Science, 289, 2000
|
|
1DIZ
 
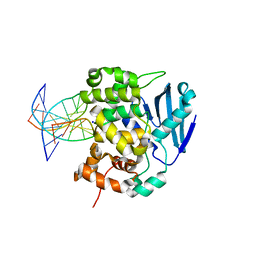 | | CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, DNA (5'-D(*GP*AP*CP*AP*TP*GP*AP*(NRI)P*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Hollis, T, Ichikawa, Y, Ellenberger, T.E. | | Deposit date: | 1999-11-30 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA.
EMBO J., 19, 2000
|
|
1DJ0
 
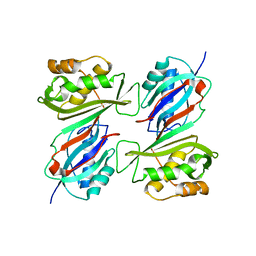 | | THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION | | Descriptor: | CHLORIDE ION, PSEUDOURIDINE SYNTHASE I | | Authors: | Foster, P.G, Huang, L, Santi, D.V, Stroud, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I.
Nat.Struct.Biol., 7, 2000
|
|
1DJ1
 
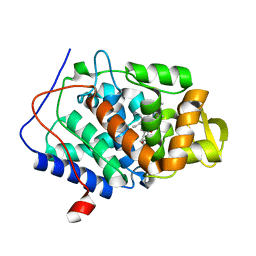 | | CRYSTAL STRUCTURE OF R48A MUTANT OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Goodin, D.B. | | Deposit date: | 1999-11-30 | | Release date: | 1999-12-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unusual oxidative chemistry of N(omega)-hydroxyarginine and N-hydroxyguanidine catalyzed at an engineered cavity in a heme peroxidase.
J.Biol.Chem., 275, 2000
|
|
1DJ2
 
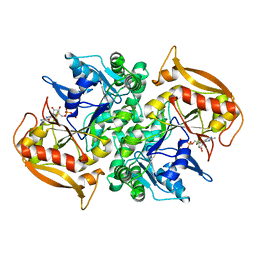 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1DJ3
 
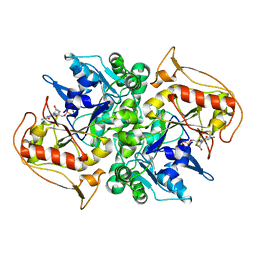 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1DJ5
 
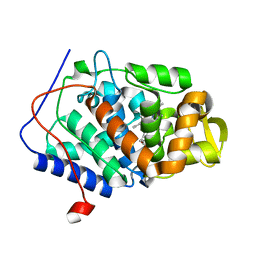 | |
1DJ6
 
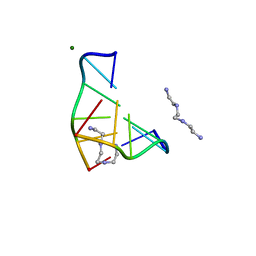 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
1DJ7
 
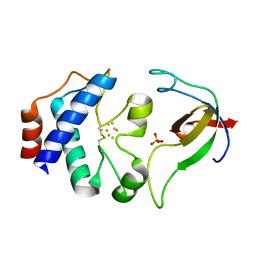 | | CRYSTAL STRUCTURE OF FERREDOXIN THIOREDOXIN REDUCTASE | | Descriptor: | FERREDOXIN THIOREDOXIN REDUCTASE: CATALYTIC CHAIN, FERREDOXIN THIOREDOXIN REDUCTASE: VARIABLE CHAIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dai, S, Schwendtmayer, C, Schurmann, P, Ramaswamy, S, Eklund, H. | | Deposit date: | 1999-12-02 | | Release date: | 2000-02-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox signaling in chloroplasts: cleavage of disulfides by an iron-sulfur cluster.
Science, 287, 2000
|
|
1DJ8
 
 | |
1DJ9
 
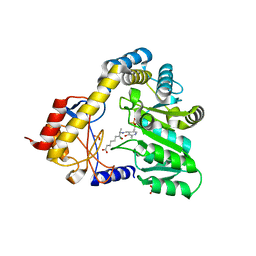 | | CRYSTAL STRUCTURE OF 8-AMINO-7-OXONANOATE SYNTHASE (OR 7-KETO-8AMINIPELARGONATE OR KAPA SYNTHASE) COMPLEXED WITH PLP AND THE PRODUCT 8(S)-AMINO-7-OXONANONOATE (OR KAPA). THE ENZYME OF BIOTIN BIOSYNTHETIC PATHWAY. | | Descriptor: | 8-AMINO-7-OXONONANOATE SYNTHASE, MAGNESIUM ION, N-[7-KETO-8-AMINOPELARGONIC ACID]-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1DJA
 
 | |
1DJB
 
 | |
1DJC
 
 | |
1DJD
 
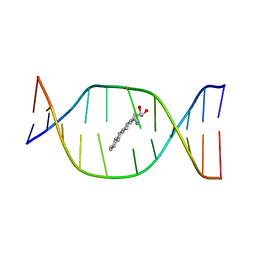 | | THE SOLUTION STRUCTURE OF A NON-BAY REGION 11R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 8,9,10,11-TETRAHYDRO-BENZO[A]ANTHRACENE-8,9,10-TRIOL, DNA(5'-D(*CT*GT*GT*AT*CT*AT*AT*GT*AT*AT*G)-3'), DNA(5'-D(*CT*TT*TT*CT*TT*TT*GT*TT*CT*CT*G)-3') | | Authors: | Li, Z, Kim, H.Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-12-02 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Role of a polycyclic aromatic hydrocarbon bay region ring in modulating DNA adduct structure: the non-bay region (8S,9R,10S, 11R)-N(6)-[11-(8,9,10,11-tetrahydro-8,9, 10-trihydroxybenz[a]anthracenyl)]-2' -deoxyadenosyl adduct in codon 61 of the human N-ras protooncogene
Biochemistry, 38, 1999
|
|
1DJE
 
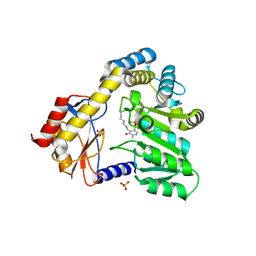 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF 8-AMINO-7-OXONANOATE SYNTHASE | | Descriptor: | 8-AMINO-7-OXONANOATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DJG
 
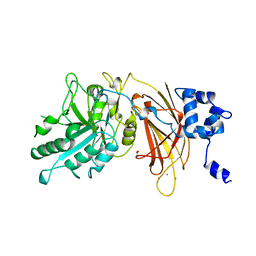 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH LANTHANUM | | Descriptor: | ACETATE ION, LANTHANUM (III) ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1DJH
 
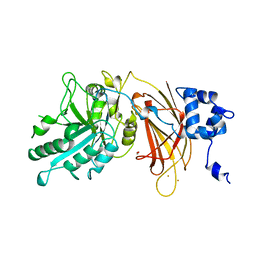 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH BARIUM | | Descriptor: | ACETATE ION, BARIUM ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
