5WSU
 
 | | Crystal structure of Myosin VIIa IQ5-SAH in complex with apo-CaM | | Descriptor: | Calmodulin, Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
5W8U
 
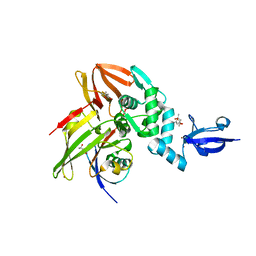 | | Crystal structure of MERS-CoV papain-like protease in complex with the C-terminal domain of human ISG15 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ORF1ab, Ubiquitin-like protein ISG15, ... | | Authors: | Daczkowski, C.M, Goodwin, O.Y, Dzimianski, J.D, Pegan, S.D. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structurally Guided Removal of DeISGylase Biochemical Activity from Papain-Like Protease Originating from Middle East Respiratory Syndrome Coronavirus.
J. Virol., 91, 2017
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W9O
 
 | |
5WTY
 
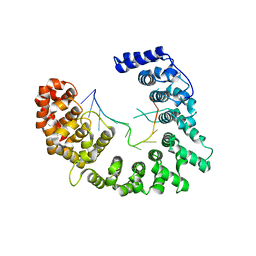 | | Structure of Nop9 RNA complex | | Descriptor: | Nucleolar protein 9, RNA (5'-R(*AP*AP*AP*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*GP*AP*AP*GP*G)-3') | | Authors: | Ye, K, Wang, B. | | Deposit date: | 2016-12-15 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Nop9 binds the central pseudoknot region of 18S rRNA
Nucleic Acids Res., 45, 2017
|
|
5WC0
 
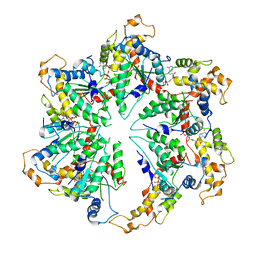 | | katanin hexamer in spiral conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Meiotic spindle formation protein mei-1 | | Authors: | Zehr, E.A, Szyk, A, Piszczek, G, Szczesna, E, Zuo, X, Roll-Mecak, A. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5WOD
 
 | |
5LGD
 
 | | The CIDRa domain from MCvar1 PfEMP1 bound to CD36 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hsieh, F.L, Higgins, M.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structural basis for CD36 binding by the malaria parasite.
Nat Commun, 7, 2016
|
|
5W7J
 
 | | X-ray structure of the E89A variant of ankyrin repeat domain of DHHC17 in complex with Snap25b peptide | | Descriptor: | Palmitoyltransferase ZDHHC17, Snap25b-111-120 | | Authors: | Verardi, R, Kim, J.-S, Ghirlando, R, Banerjee, A. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.
Structure, 25, 2017
|
|
5W7N
 
 | |
5LUB
 
 | | Crystal structure of human legumain (AEP) in complex with compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(morpholin-4-yl)-2,1,3-benzoxadiazol-4-amine, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
5W85
 
 | | CRYSTAL STRUCTURE OF IRAK-4 WITH A 4,6-DIAMINONICOTINAMIDE INHIBITOR (COMPOUND NUMBER 9) | | Descriptor: | 6-[(1,3-benzothiazol-6-yl)amino]-4-{[(2S)-1-hydroxy-3-phenylpropan-2-yl]amino}-N-methylpyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Sack, J.S. | | Deposit date: | 2017-06-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and structure-based design of 4,6-diaminonicotinamides as potent and selective IRAK4 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5W8T
 
 | | Crystal structure of MERS-CoV papain-like protease in complex with the C-terminal domain of human ISG15 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ORF1ab, Ubiquitin-like protein ISG15, ... | | Authors: | Daczkowski, C.M, Goodwin, O.Y, Dzimianski, J.V, Farhat, J.J, Pegan, S.D. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Structurally Guided Removal of DeISGylase Biochemical Activity from Papain-Like Protease Originating from Middle East Respiratory Syndrome Coronavirus.
J. Virol., 91, 2017
|
|
6DTQ
 
 | | Maltose bound T. maritima MalE3 | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
5WDQ
 
 | | H-Ras mutant L120A bound to GMP-PNP at 100K | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5W9M
 
 | |
5WK2
 
 | | CRYSTAL STRUCTURE OF ANTI-CCL17 ANTIBODY M116 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, M116 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into chemokine CCL17 recognition by antibody M116.
Biochem Biophys Rep, 13, 2018
|
|
5WEM
 
 | | GluA2 bound to GSG1L in digitonin, state 1 | | Descriptor: | Chimera of Glutamate receptor 2,Germ cell-specific gene 1-like protein | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
5WKO
 
 | |
5WMA
 
 | | N-terminal bromodomain of BRD4 in complex with PLX5981 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMU
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase I | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WN8
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lewis-Ballester, A, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L, Yeh, S.R. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WGV
 
 | | Crystal Structure of MalA' C112S/C128S, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WH6
 
 | | Crystal structure of PDE4D2 in complex with inhibitor (S_Zl-n-91) | | Descriptor: | 1-[4-(difluoromethoxy)-3-{[(3S)-oxolan-3-yl]oxy}phenyl]-3-methylbutan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H, Wang, H. | | Deposit date: | 2017-07-14 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a PDE4-Specific Pocket for the Design of Selective Inhibitors.
Biochemistry, 57, 2018
|
|
5WJ0
 
 | | Phosphotriesterase variant S5+254R | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphotriesterase variant S5+254R
To Be Published
|
|
