8Y3C
 
 | | Cryo-EM structure of the overlapping di-nucleosome (closed form) | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tanaka, H, Nishimura, M, Takizawa, Y, Nozawa, K, Ehara, H, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-01-29 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (5.21 Å) | | Cite: | Asymmetric fluctuation of overlapping dinucleosome studied by cryoelectron microscopy and small-angle X-ray scattering.
Pnas Nexus, 3, 2024
|
|
8Y3E
 
 | | Cryo-EM structure of the overlapping di-nucleosome (open form) | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tanaka, H, Nishimura, M, Takizawa, Y, Nozawa, K, Ehara, H, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-01-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Asymmetric fluctuation of overlapping dinucleosome studied by cryoelectron microscopy and small-angle X-ray scattering.
Pnas Nexus, 3, 2024
|
|
8XRJ
 
 | | RNA polymerase II elongation complex with upstream nucleosome extracted from human nuclei | | Descriptor: | DNA, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Kujirai, T, Kato, J, Yamamoto, K, Hirai, S, Negishi, L, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-01-07 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Multiple structures of RNA polymerase II isolated from human nuclei by ChIP-CryoEM analysis.
Nat Commun, 16, 2025
|
|
8XVS
 
 | | RNA polymerase II elongation complex with downstream nucleosome extracted from human nuclei | | Descriptor: | DNA, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Kujirai, T, Kato, J, Yamamoto, K, Hirai, S, Negishi, L, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-01-15 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Multiple structures of RNA polymerase II isolated from human nuclei by ChIP-CryoEM analysis.
Nat Commun, 16, 2025
|
|
8YJM
 
 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and a single chain H2B-H2A chimera | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2B 1/2/3/4/6,Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
8YTI
 
 | | Crystal Structure of Nucleosome-H1x Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Qiuye, B, Padavattan, S, Davey, C.A. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Linker Histones Associate Heterogeneously with Nucleosomes in the Condensed State
To Be Published
|
|
8YJF
 
 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and an H2A-H2B dimer | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
8Z50
 
 | | Crystal structure of the ASF1-H3T-H4 complex | | Descriptor: | Histone H3.1t, Histone H4, Histone chaperone ASF1A | | Authors: | Xu, L. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into instability of the nucleosome driven by histone variant H3T.
Biochem.Biophys.Res.Commun., 727, 2024
|
|
8YV8
 
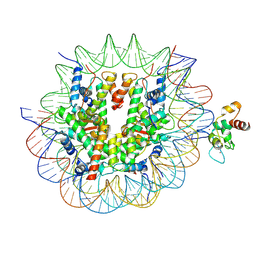 | | Cryo-EM structure of CDCA7 bound to nucleosome including hemimethylated CpG site in Widom601 positioning sequence. | | Descriptor: | Cell division cycle-associated protein 7, DNA (132-MER), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, A, Shikimachi, R, Nishiyama, A, Funabiki, H, Arita, K. | | Deposit date: | 2024-03-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CDCA7 is an evolutionarily conserved hemimethylated DNA sensor in eukaryotes.
Sci Adv, 10, 2024
|
|
8YNY
 
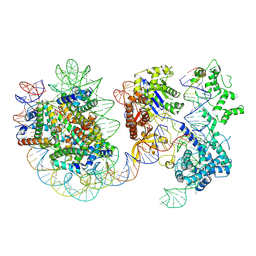 | | Structure of Cas9-sgRNA ribonucleoprotein bound to nucleosome | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (17-mer), DNA (175-mer), ... | | Authors: | Nagamura, R, Kujirai, T, Kusakizako, T, Hirano, H, Kurumizaka, H, Nureki, O. | | Deposit date: | 2024-03-12 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Structural insights into how Cas9 targets nucleosomes.
Nat Commun, 15, 2024
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
8YBK
 
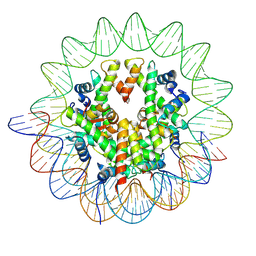 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBJ
 
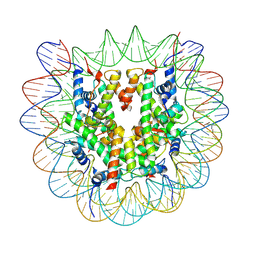 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
7PJ1
 
 | |
9GD0
 
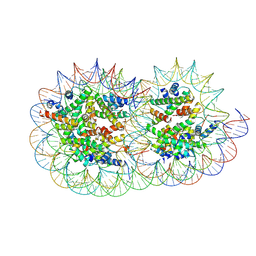 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD2
 
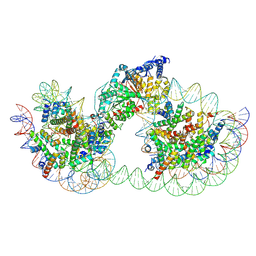 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD1
 
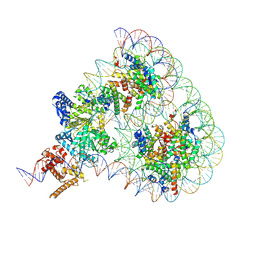 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD3
 
 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
7PFV
 
 | |
9GMR
 
 | | SIRT7-H3K36MTUnucleosome complex | | Descriptor: | DNA (149-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | Authors: | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
9GXA
 
 | | CENP-A/H4 di-tetrasome assembled on alpha-satellite DNA. | | Descriptor: | 147 bp alpha-satellite DNA, 147 bp human alpha-satellite DNA, Histone H3-like centromeric protein A, ... | | Authors: | Ali-Ahmad, A, Sekulic, N. | | Deposit date: | 2024-09-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Non-nucleosomal (CENP-A/H4) 2 - DNA complexes as a possible platform for centromere organization.
Biorxiv, 2025
|
|
9GMK
 
 | | SIRT7:H3K18DTU nucleosome complex | | Descriptor: | DNA (148-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | Authors: | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
9IPU
 
 | | cryo-EM structure of the RNF168(1-193)/UbcH5c-Ub ubiquitylation module bound to H1.0-K63-Ub3 modified chromatosome | | Descriptor: | DNA (171-MER), E3 ubiquitin-protein ligase RNF168, Histone H1.0, ... | | Authors: | Ai, H.S, Deng, Z.H, Liu, L. | | Deposit date: | 2024-07-11 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Promotion of RNF168-Mediated Nucleosomal H2A Ubiquitylation by Structurally Defined K63-Polyubiquitylated Linker Histone H1.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9II7
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome containing histone variant H2A.B | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Kujirai, T, Rina, H, Ehara, H, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-06-19 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of RNAPII transcription on the nucleosome containing histone variant H2A.B.
Embo J., 2025
|
|
7PII
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, DNA (122-MER), DNA (123-MER), ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Barford, D. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
