5HXZ
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
7N5Z
 
 | | SARS-CoV-2 Main protease C145S mutant | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
5HPR
 
 | | Insulin with proline analog HyP at position B28 in the T2 state | | Descriptor: | GLYCEROL, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
7NJ9
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-133 | | Descriptor: | 14-3-3 protein sigma, 4-(3,4-dihydro-2~{H}-quinolin-1-ylsulfonyl)benzaldehyde, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NJ6
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1005 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(trifluoromethyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NJB
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-132 | | Descriptor: | 14-3-3 protein sigma, 4-piperidin-1-ylsulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7N6N
 
 | | SARS-CoV-2 Main protease C145S mutant in complex with N and C-terminal residues | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7N50
 
 | |
7NJ8
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1007 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5HTS
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - D295N mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5HVI
 
 | |
7NQP
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound LvD1009 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-03-02 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NIX
 
 | | 14-3-3 sigma with AS160 binding site pT642 | | Descriptor: | 14-3-3 protein sigma, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7N52
 
 | |
5HOC
 
 | | p73 homo-tetramerization domain mutant II | | Descriptor: | Tumor protein p73 | | Authors: | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Sumyk, M, Knapp, S, Dotsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36007786 Å) | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
5HOG
 
 | |
7NIG
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1008 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5HPE
 
 | | Phosphatase domain of PP5 bound to a phosphomimetic Cdc37 substrate peptide | | Descriptor: | COBALT HEXAMMINE(III), MANGANESE (II) ION, Serine/threonine-protein phosphatase 5,Hsp90 co-chaperone Cdc37 | | Authors: | Oberoi, J, Mariotti, L, Vaughan, C. | | Deposit date: | 2016-01-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and functional basis of protein phosphatase 5 substrate specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7NEI
 
 | |
5HYM
 
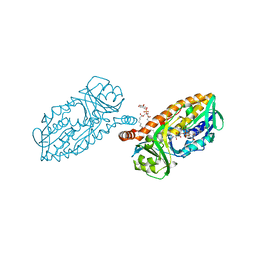 | | 3-Hydroxybenzoate 6-hydroxylase from Rhodococcus jostii in complex with phosphatidylinositol | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Phosphatidylinositol, ... | | Authors: | Orru, R, Montersino, S, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxybenzoate 6-Hydroxylase from Rhodococcus jostii RHA1 Contains a Phosphatidylinositol Cofactor.
Front Microbiol, 8, 2017
|
|
7N89
 
 | |
7N51
 
 | |
7NJA
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1006 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5HRV
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-ethylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
7NLY
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-Chlorobenzimidazole. | | Descriptor: | 2-chloranyl-1~{H}-benzimidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
