2J5C
 
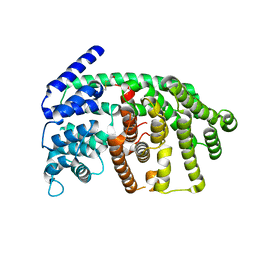 | | Rational conversion of substrate and product specificity in a monoterpene synthase. Structural insights into the molecular basis of rapid evolution. | | Descriptor: | 1,8-CINEOLE SYNTHASE, BETA-MERCAPTOETHANOL | | Authors: | Kampranis, S.C, Ioannidis, D, Purvis, A, Mahrez, W, Ninga, E, Katerelos, N.A, Anssour, S, Dunwell, J.M, Makris, A.M, Goodenough, P.W, Johnson, C.B. | | Deposit date: | 2006-09-14 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Conversion of Substrate and Product Specificity in a Salvia Monoterpene Synthase: Structural Insights Into the Evolution of Terpene Synthase Function.
Plant Cell, 19, 2007
|
|
2J5D
 
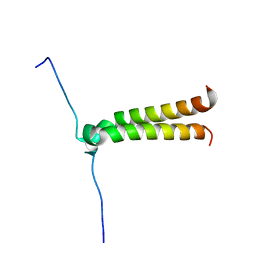 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
2J5E
 
 | |
2J5F
 
 | |
2J5G
 
 | | The Native structure of a beta-Diketone Hydrolase from the Cyanobacterium Anabaena sp. PCC 7120 | | Descriptor: | ALR4455 PROTEIN, SULFATE ION | | Authors: | Bennett, J.P, Whittingham, J.L, Brzozowski, A.M, Leonard, P.M, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Characterisation of a Beta Diketone Hydrolase from the Cyanobacterium Anabaena Sp. Pcc 7120 in Native and Product Bound Forms, a Coenzyme A-Independent Member of the Crotonase Suprafamily
Biochemistry, 46, 2007
|
|
2J5H
 
 | | NMR analysis of mouse CRIPTO CFC domain | | Descriptor: | TERATOCARCINOMA-DERIVED GROWTH FACTOR | | Authors: | Calvanese, L, Saporito, A, Marasco, D, D'Auria, G, Minchiotti, G, Pedone, C, Paolillo, L, Falcigno, L, Ruvo, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse Cripto CFC domain and its inactive variant Trp107Ala.
J. Med. Chem., 49, 2006
|
|
2J5I
 
 | | Crystal Structure of Hydroxycinnamoyl-CoA Hydratase-Lyase | | Descriptor: | P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Leonard, P.M, Brzozowski, A.M, Lebedev, A, Marshall, C.M, Smith, D.J, Verma, C.S, Walton, N.J, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5L
 
 | | Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
2J5M
 
 | | Structure of Chloroperoxidase Compound 0 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuhnel, K, Derat, E, Terner, J, Shaik, S, Schlichting, I. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Quantum Chemical Characterization of Chloroperoxidase Compound 0, a Common Reaction Intermediate of Diverse Heme Enzymes.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2J5N
 
 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE FROM THERMUS THERMOPHIRUS WITH BOUND INHIBITOR GLYCINE AND NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of Ternary Complex of Delta1-Pyrroline-5-Carboxylate Dehydrogenase with Substrate Mimic and Co-Factoer
To be Published
|
|
2J5O
 
 | | Pseudomonas aeruginosa FtsK gamma domain | | Descriptor: | DNA TRANSLOCASE FTSK | | Authors: | Sivanathan, V, Allen, M.D, deBekker, C, Baker, R, Arciszewska, L, Freund, S.M, Bycroft, M, Lowe, J, Sherratt, D.J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ftsk Gamma Domain Directs Oriented DNA Translocation by Interacting with Kops.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2J5P
 
 | | E. coli FtsK gamma domain | | Descriptor: | DNA TRANSLOCASE FTSK | | Authors: | Sivanathan, V, Allen, M.D, de Bekker, C, Baker, R, Arciszewska, L, Freund, S.M, Bycroft, M, Lowe, J, Sherratt, D.J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ftsk Gamma Domain Directs Oriented DNA Translocation by Interacting with Kops.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2J5Q
 
 | | 2.15 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after first radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5R
 
 | | 2.25 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after second radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5S
 
 | | Structural of ABDH, a beta-diketone hydrolase from the Cyanobacterium Anabaena sp. PCC 7120 bound to (S)-3-oxocyclohexyl acetic acid | | Descriptor: | (S)-CYCLOHEXANONE-2-ACETATE, BETA-DIKETONE HYDROLASE, NICKEL (II) ION | | Authors: | Bennett, J.P, Whittingham, J.L, Brzozowski, A.M, Leonard, P.M, Grogan, G. | | Deposit date: | 2006-09-19 | | Release date: | 2007-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterisation of a Beta Diketone Hydrolase from the Cyanobacterium Anabaena Sp. Pcc 7120 in Native and Product Bound Forms, a Coenzyme A-Independent Member of the Crotonase Suprafamily
Biochemistry, 46, 2007
|
|
2J5T
 
 | | Glutamate 5-kinase from Escherichia coli complexed with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE 5-KINASE, GLUTAMIC ACID, ... | | Authors: | Marco-Marin, C, Gil-Ortiz, F, Perez-Arellano, I, Cervera, J, Fita, I, Rubio, V. | | Deposit date: | 2006-09-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Two-Domain Architecture within the Amino Acid Kinase Enzyme Family Revealed by the Crystal Structure of Escherichia Coli Glutamate 5-Kinase.
J.Mol.Biol., 367, 2007
|
|
2J5U
 
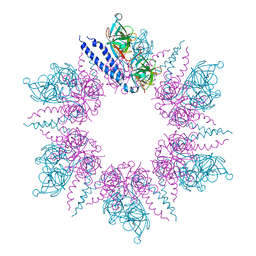 | | MreC Lysteria monocytogenes | | Descriptor: | MREC PROTEIN | | Authors: | van den Ent, F, Leaver, M, Bendezu, F, Errington, J, de Boer, P, Lowe, J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric Structure of the Cell Shape Protein Mrec and its Functional Implications.
Mol.Microbiol., 62, 2006
|
|
2J5V
 
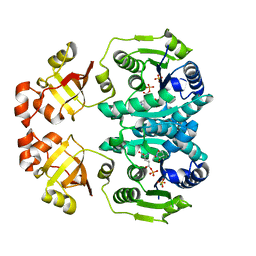 | | GLUTAMATE 5-KINASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTAMYL-5-PHOSPHATE AND PYROGLUTAMIC ACID | | Descriptor: | GAMMA-GLUTAMYL PHOSPHATE, GLUTAMATE 5-KINASE, MAGNESIUM ION, ... | | Authors: | Marco-Marin, C, Gil-Ortiz, F, Perez-Arellano, I, Cervera, J, Fita, I, Rubio, V. | | Deposit date: | 2006-09-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Two-Domain Architecture within the Amino Acid Kinase Enzyme Family Revealed by the Crystal Structure of Escherichia Coli Glutamate 5-Kinase.
J.Mol.Biol., 367, 2007
|
|
2J5W
 
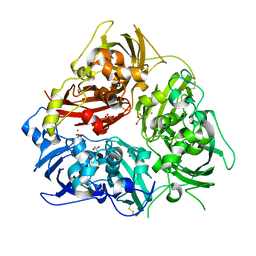 | | Ceruloplasmin revisited: structural and functional roles of various metal cation binding sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CERULOPLASMIN, ... | | Authors: | Bento, I, Peixoto, C, Zaitsev, V.N, Lindley, P.F. | | Deposit date: | 2006-09-19 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ceruloplasmin Revisited: Structural and Functional Roles of Various Metal Cation-Binding Sites.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2J5X
 
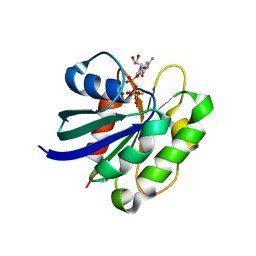 | | STRUCTURE OF THE SMALL G PROTEIN ARF6 IN COMPLEX WITH GTPGAMMAS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-RIBOSYLATION FACTOR 6, MAGNESIUM ION | | Authors: | Pasqualato, S, Menetrey, J, Franco, M, Cherfils, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structural Gdp-GTP Cycle of Human Arf6.
Embo Rep., 2, 2001
|
|
2J5Y
 
 | | Crystal structure of the GA module from F.magna | | Descriptor: | PEPTOSTREPTOCOCCAL ALBUMIN-BINDING PROTEIN | | Authors: | Lejon, S, Cramer, J.F, Nordberg, P.A, Lundqvist, T, Valegard, K. | | Deposit date: | 2006-09-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Bacterial Albumin-Binding Domain at 1.4A Resolution.
FEBS Lett., 581, 2007
|
|
2J5Z
 
 | | H-ficolin complexed to galactose | | Descriptor: | ACETATE ION, CALCIUM ION, FICOLIN-3, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-21 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J60
 
 | | H-ficolin complexed to D-fucose | | Descriptor: | ACETATE ION, CALCIUM ION, FICOLIN-3, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-21 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J61
 
 | |
