2IJD
 
 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
2IJE
 
 | |
2IJF
 
 | |
2IJG
 
 | | Crystal Structure of cryptochrome 3 from Arabidopsis thaliana | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, Cryptochrome DASH, chloroplast/mitochondrial, ... | | Authors: | Huang, Y, Deisenhofer, J. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2IJH
 
 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJI
 
 | | Structure of F14H mutant of ColE1 Rom protein | | Descriptor: | Regulatory protein rop | | Authors: | Struble, E.B. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJJ
 
 | | Crystal structure analysis of ColE1 ROM mutant F14Y | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJK
 
 | | Structure of a Rom protein dimer at 1.55 angstrom resolution | | Descriptor: | Regulatory protein rop | | Authors: | Struble, E.B. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJL
 
 | | The structure of a putative ModE from Agrobacterium tumefaciens. | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum-binding transcriptional repressor, SULFATE ION | | Authors: | Cuff, M.E, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative ModE from Agrobacterium tumefaciens.
To be Published
|
|
2IJM
 
 | |
2IJN
 
 | | Isothiazoles as active-site inhibitors of HCV NS5B polymerase | | Descriptor: | (2R,3R)-3-{[3,5-BIS(TRIFLUOROMETHYL)PHENYL]AMINO}-2-CYANO-3-THIOXOPROPANAMIDE, RNA polymerase NS5B | | Authors: | Yan, S, Yao, N. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazoles as active-site inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2IJO
 
 | | Crystal Structure of the West Nile virus NS2B-NS3 protease complexed with bovine pancreatic trypsin inhibitor | | Descriptor: | Pancreatic trypsin inhibitor, Polyprotein | | Authors: | Aleshin, A.E, Shiryaev, S.A, Strongin, A.Y, Liddington, R.C. | | Deposit date: | 2006-09-29 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold.
Protein Sci., 16, 2007
|
|
2IJQ
 
 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
2IJR
 
 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
2IJX
 
 | |
2IJY
 
 | |
2IJZ
 
 | |
2IK0
 
 | | Yeast inorganic pyrophosphatase variant E48D with magnesium and phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK1
 
 | | Yeast inorganic pyrophosphatase variant Y93F with magnesium and phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK2
 
 | | Yeast inorganic pyrophosphatase variant D115E with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK4
 
 | | Yeast inorganic pyrophosphatase variant D117E with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK6
 
 | | Yeast inorganic pyrophosphatase variant D120E with magnesium and phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK7
 
 | | Yeast inorganic pyrophosphatase variant D120N with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK8
 
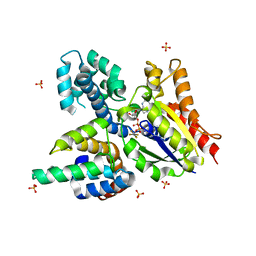 | | Crystal structure of the heterodimeric complex of human RGS16 and activated Gi alpha 1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Soundararajan, M, Turnbull, A.P, Papagrigoriou, E, Debreczeni, J, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-02 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2IK9
 
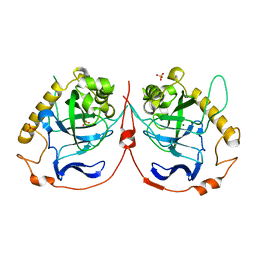 | | Yeast inorganic pyrophosphatase variant D152E with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
