3D5Z
 
 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (AbnBE201A) complexed to arabinotriose | | Descriptor: | CALCIUM ION, Intracellular arabinanase, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-beta-L-arabinofuranose | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-05-18 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
6CUM
 
 | |
7AKM
 
 | | Crystal structure of CHK1 kinase domain in complex with ATPyS | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for recruitment of the CHK1 DNA damage kinase by the CLASPIN scaffold protein.
Structure, 29, 2021
|
|
6HPV
 
 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
4G3F
 
 | |
3D1K
 
 | | R/T intermediate quaternary structure of an antarctic fish hemoglobin in an alpha(CO)-beta(pentacoordinate) state | | Descriptor: | ACETYL GROUP, CARBON MONOXIDE, Hemoglobin subunit alpha-1, ... | | Authors: | Vitagliano, L, Vergara, A, Bonomi, G, Merlino, A, Mazzarella, L. | | Deposit date: | 2008-05-06 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopic and crystallographic characterization of a tetrameric hemoglobin oxidation reveals structural features of the functional intermediate relaxed/tense state.
J.Am.Chem.Soc., 130, 2008
|
|
9F63
 
 | |
3LZM
 
 | |
8X91
 
 | | P/Q type calcium channel in complex with omega-conotoxin MVIIC | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
7ZMK
 
 | |
6W6G
 
 | |
6W6H
 
 | |
6W6E
 
 | |
6W6J
 
 | |
6W6I
 
 | |
6WGC
 
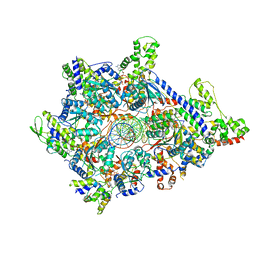 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8I0J
 
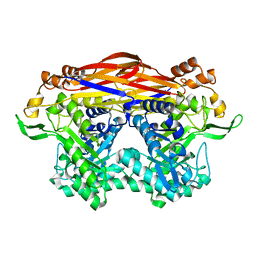 | | JB13GH39P28 mutant-D41G | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | beta-Xylosidase JB13GH39P28 (D41G) showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
7O0O
 
 | | Crystal structure of the B3 metallo-beta-lactamase L1 with hydrolysed ertapenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallography and QM/MM Simulations Identify Preferential Binding of Hydrolyzed Carbapenem and Penem Antibiotics to the L1 Metallo-beta-Lactamase in the Imine Form.
J.Chem.Inf.Model., 61, 2021
|
|
3VEG
 
 | | Rhodococcus jostii RHA1 DypB R244L variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3VED
 
 | | Rhodococcus jostii RHA1 DypB D153H variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, GLYCEROL, ... | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3VEE
 
 | | Rhodococcus jostii RHA1 DypB N246A variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, FORMIC ACID, ... | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
8BSU
 
 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate and the positive allosteric modulator BPAM344 at 2.9A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
8BST
 
 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate at 2.7A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
8A4F
 
 | | Human Interleukin-4 mutant - C3T-IL4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Mueller, T.D, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2022-06-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
6FER
 
 | | Crystal Structure of human DDR2 kinase in complex with 2-[4,5-difluoro-2-oxo-1'-(1H-pyrazolo[3,4-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide | | Descriptor: | 2-[4,5-bis(fluoranyl)-2-oxidanylidene-1'-(1~{H}-pyrazolo[3,4-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Discoidin domain-containing receptor 2 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
