7NKA
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8206 | | Descriptor: | NB8206, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIL
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8190 core | | Descriptor: | Nanobody8190 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8191 core | | Descriptor: | Nanobody8191 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK2
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8202 core | | Descriptor: | Nanobody8202, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ4
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8198 core | | Descriptor: | Nb8198 Core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK4
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8203 core | | Descriptor: | Nanobody 8203, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ3
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8196 core | | Descriptor: | Nanobody8196 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK8
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8205 core | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7CUN
 
 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
2F3I
 
 | | Solution Structure of a Subunit of RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerases I, II, and III 17.1 kDa polypeptide | | Authors: | Kang, X, Jin, C. | | Deposit date: | 2005-11-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, biochemical, and dynamic characterizations of the hRPB8 subunit of human RNA polymerases
J.Biol.Chem., 281, 2006
|
|
6IS0
 
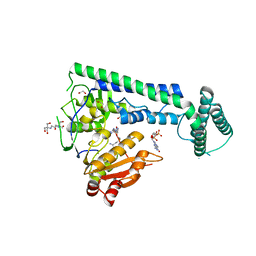 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
4Q5S
 
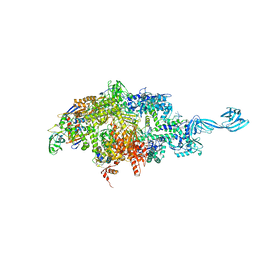 | | Thermus thermophilus RNA polymerase initially transcribing complex containing 6-mer RNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*GP*CP*AP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3'), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
1QKL
 
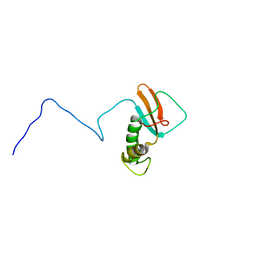 | | hRPABC14.4, essential subunit of human RNA polymerases I, II and III | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 14.4 KD POLYPEPTIDE | | Authors: | Del Rio-Portilla, F, Gaskell, A.G, Gilbert, D, Ladias, J.A.A, Wagner, G. | | Deposit date: | 1999-07-26 | | Release date: | 1999-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Hrpabc14.4 Subunit of Human RNA Polymerases
Nat.Struct.Biol., 6, 1999
|
|
4H3K
 
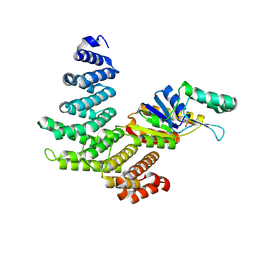 | | Crystal structure of a ternary complex of human symplekin NTD, human Ssu72 and a RNA polymerase II CTD peptide phosphorylated at Ser-2, Ser-5 and Ser-7 | | Descriptor: | Hexapeptide, PHOSPHATE ION, RNA polymerase II subunit A C-terminal domain phosphatase SSU72, ... | | Authors: | Xiang, K, Tong, L. | | Deposit date: | 2012-09-13 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72.
Genes Dev., 26, 2012
|
|
1Y14
 
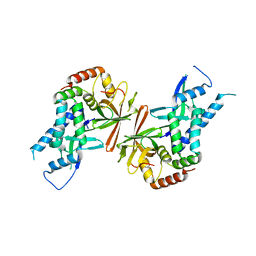 | | Crystal structure of yeast subcomplex of Rpb4 and Rpb7 | | Descriptor: | DNA-directed RNA polymerase II 19 kDa polypeptide, DNA-directed RNA polymerase II 32 kDa polypeptide | | Authors: | Armache, K.-J, Mitterweger, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Complete RNA Polymerase II and Its Subcomplex, Rpb4/7
J.Biol.Chem., 280, 2005
|
|
1SZA
 
 | |
1SZ9
 
 | |
2L0I
 
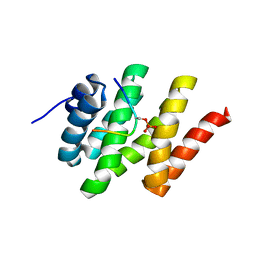 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4FQG
 
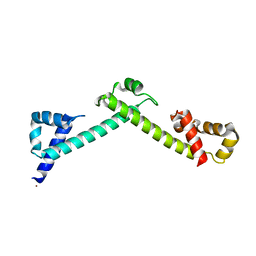 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | Authors: | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
6EU3
 
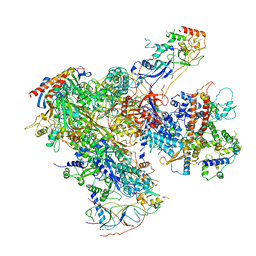 | | Apo RNA Polymerase III - closed conformation (cPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
8QQ2
 
 | |
8QW9
 
 | |
7NKY
 
 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5OA1
 
 | | RNA polymerase I pre-initiation complex | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Sadian, Y, Tafur, L, Kosinski, J, Jakobi, A.J, Muller, C.W. | | Deposit date: | 2017-06-20 | | Release date: | 2017-07-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into transcription initiation by yeast RNA polymerase I.
EMBO J., 36, 2017
|
|
5N61
 
 | | RNA polymerase I initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
