6HZJ
 
 | | Apo structure of TP domain from clinical penicillin-resistant mutant Neisseria gonorrhoea strain 6140 Penicillin-Binding Protein 2 (PBP2) | | Descriptor: | Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZO
 
 | | Apo structure of TP domain from Haemophilus influenzae Penicillin-Binding Protein 3 | | Descriptor: | FtsI | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZQ
 
 | | Apo structure of TP domain from Escherichia coli Penicillin-Binding Protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZI
 
 | |
6HZR
 
 | | Apo structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Bellini, D, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6I1I
 
 | | Crystal structure of TP domain from Escherichia coli penicillin-binding protein 3 in complex with penicillin | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI,Peptidoglycan D,D-transpeptidase FtsI, Piperacillin (Open Form) | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6Q9N
 
 | | Crystal structure of PBP2a from MRSA in complex with piperacillin and quinazolinone | | Descriptor: | 3-[2-[(~{E})-2-(4-ethynylphenyl)ethenyl]-4-oxidanylidene-quinazolin-3-yl]benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Martinez-Caballero, S, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus.
Antimicrob.Agents Chemother., 63, 2019
|
|
6V6N
 
 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | Descriptor: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-05 | | Release date: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6PK0
 
 | | Crystal Structure of OXA-48 with Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
6PQI
 
 | |
6PSG
 
 | | Crystal Structure of Class D Beta-lactamase OXA-48 with Faropenem | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, CHLORIDE ION, Class D Carbapenemase OXA-48, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
6PTU
 
 | |
6PT5
 
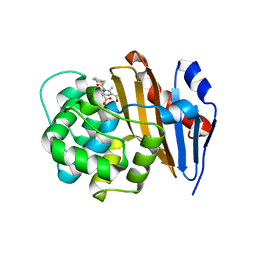 | | Crystal Structure of Class D Beta-lactamase OXA-48 with Cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Class D Carbapenemase OXA-48 | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-07-14 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
6PT1
 
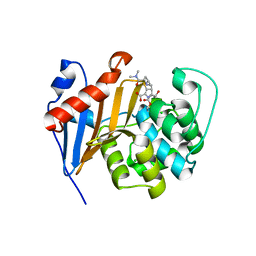 | | Crystal Structure of Class D Beta-lactamase OXA-48 with Meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, CHLORIDE ION, Class D Carbapenemase OXA-48, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-07-14 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
6NZ8
 
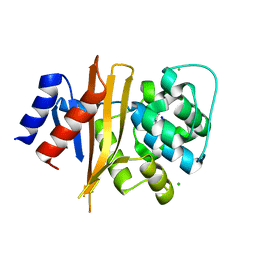 | | Structure of carbamylated apo OXA-231 carbapenemase | | Descriptor: | Beta-lactamase OXA-231, CHLORIDE ION, SODIUM ION | | Authors: | Favaro, D.C, Llontop, E.E, Vasconcelos, F.N, Antunes, V.U, Farah, S.C, Lincopan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Biochemistry, 58, 2019
|
|
6U58
 
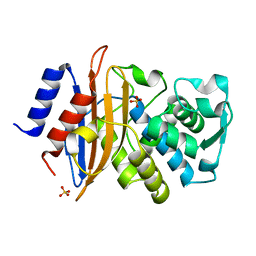 | | Toho1 Beta Lactamase Glu166Gln Mutant | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Langan, P.S, Sullivan, B, Weiss, K.L. | | Deposit date: | 2019-08-27 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Probing the Role of the Conserved Residue Glu166 in a Class A Beta-Lactamase Using Neutron and X-ray Protein Crystallography
Acta Crystallogr.,Sect.D, 76, 2020
|
|
6R40
 
 | |
6R3X
 
 | |
6R42
 
 | |
6I5D
 
 | | Crystal structure of an OXA-48 beta-lactamase synthetic mutant | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Zavala, A, Retailleau, P, Dabos, L, Naas, T, Iorga, B. | | Deposit date: | 2018-11-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate specificity of an OXA-48 beta-lactamase synthetic mutant
To be published
|
|
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGT
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
