3GMO
 
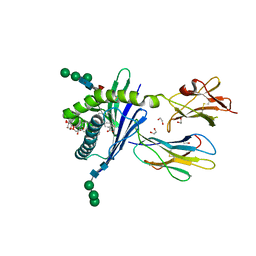 | | Structure of mouse CD1d in complex with C8PhF | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-(4-fluorophenyl)-N-{(1S,2S,3R)-1-[(alpha-D-galactopyranosyloxy)methyl]-2,3-dihydroxyheptadecyl}octanamide, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
2YNM
 
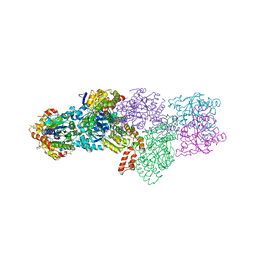 | | Structure of the ADPxAlF3-Stabilized Transition State of the Nitrogenase-like Dark-Operative Protochlorophyllide Oxidoreductase Complex from Prochlorococcus marinus with Its Substrate Protochlorophyllide a | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Krausze, J, Lange, C, Heinz, D.W, Moser, J. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Adp-Aluminium Fluoride-Stabilized Protochlorophyllide Oxidoreductase Complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7X2S
 
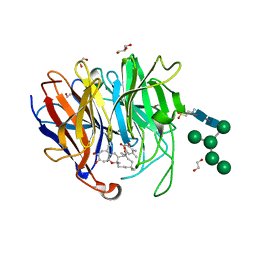 | | Crystal Structure of hetero-Diels-Alderase PycR1 in complex with Neosetophomone B and tropolone o-quinone methide | | Descriptor: | (1~{S},3~{R},11~{R},13~{S},14~{Z},18~{E})-1,8,14,17,17-pentamethyl-5,13-bis(oxidanyl)-2-oxatricyclo[9.9.0.0^{3,9}]icosa-4,7,9,14,18-pentaen-6-one, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
4IDM
 
 | |
1HDZ
 
 | |
2INQ
 
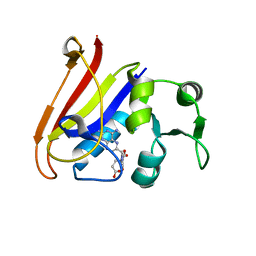 | | Neutron Crystal Structure of Escherichia coli Dihydrofolate Reductase Bound to the Anti-cancer drug, Methotrexate | | Descriptor: | Dihydrofolate reductase, N-(4-{[(2,4-DIAMINOPTERIDIN-1-IUM-6-YL)METHYL](METHYL)AMINO}BENZOYL)-L-GLUTAMIC ACID | | Authors: | Bennett, B.C, Langan, P.A, Coates, L, Schoenborn, B, Dealwis, C.G. | | Deposit date: | 2006-10-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1DB1
 
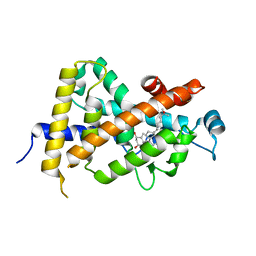 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | Authors: | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
1DGH
 
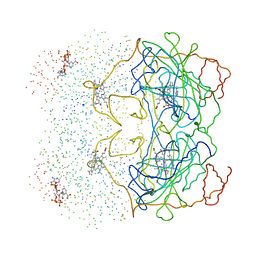 | | HUMAN ERYTHROCYTE CATALASE 3-AMINO-1,2,4-TRIAZOLE COMPLEX | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (CATALASE), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Putnam, C.D, Arvai, A.S, Bourne, Y, Tainer, J.A. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism.
J.Mol.Biol., 296, 2000
|
|
4J3Q
 
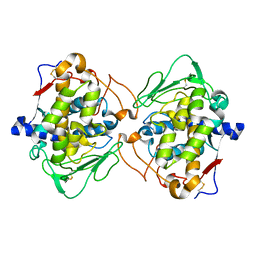 | | Crystal structure of truncated catechol oxidase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Hakulinen, N, Gasparetti, C, Kaljunen, H, Rouvinen, J. | | Deposit date: | 2013-02-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae.
J.Biol.Inorg.Chem., 18, 2013
|
|
1DFG
 
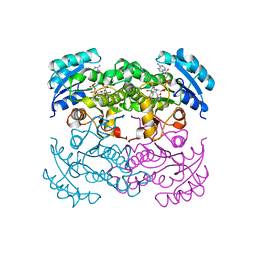 | | X-RAY STRUCTURE OF ESCHERICHIA COLI ENOYL REDUCTASE WITH BOUND NAD AND BENZO-DIAZABORINE | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ENOYL ACYL CARRIER PROTEIN REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baldock, C, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanism of drug action revealed by structural studies of enoyl reductase.
Science, 274, 1996
|
|
4F3I
 
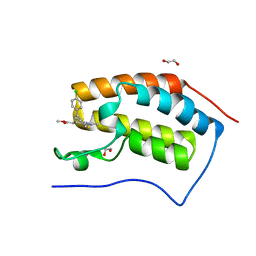 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Joshua, J, Zhou, M.-M, Plotnikov, A.N. | | Deposit date: | 2012-05-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Down-regulation of NF-{kappa}B Transcriptional Activity in HIV-associated Kidney Disease by BRD4 Inhibition.
J.Biol.Chem., 287, 2012
|
|
3VKZ
 
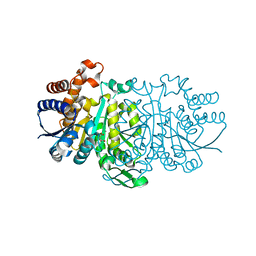 | |
5O9S
 
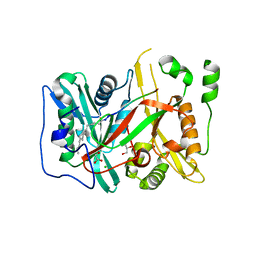 | | HsNMT1 in complex with CoA and Myristoylated-GKSNSKLK octapeptide | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Meinnel, T, Giglione, C. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
1DZN
 
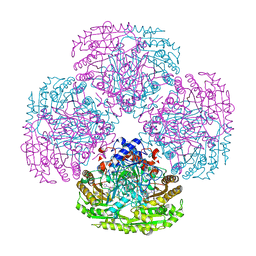 | | Asp170Ser mutant of vanillyl-alcohol oxidase | | Descriptor: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, FLAVIN-ADENINE DINUCLEOTIDE, VANILLYL-ALCOHOL OXIDASE | | Authors: | Van Den heuvel, R.H.H, Fraaije, M.W, Van Berkel, W.J.H, Mattevi, A. | | Deposit date: | 2000-03-05 | | Release date: | 2000-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asp170 is Crucial for the Redox Properties of Vanillyl-Alcohol Oxidase
J.Biol.Chem., 275, 2000
|
|
1HKX
 
 | | Crystal structure of calcium/calmodulin-dependent protein kinase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II ALPHA CHAIN, CHLORIDE ION, ... | | Authors: | Hoelz, A, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2003-03-12 | | Release date: | 2003-06-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of a Tetradecameric Assembly of the Association Domain of Ca2+/Calmodulin-Dependent Kinase II
Mol.Cell, 11, 2003
|
|
6EG0
 
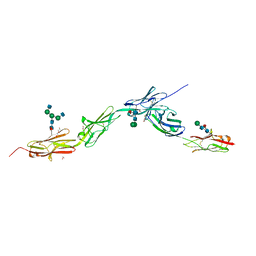 | | Crystal structure of Dpr4 Ig1-Ig2 in complex with DIP-Eta Ig1-Ig3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
1HJ4
 
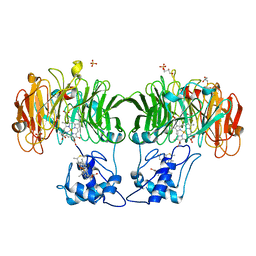 | |
1HJ3
 
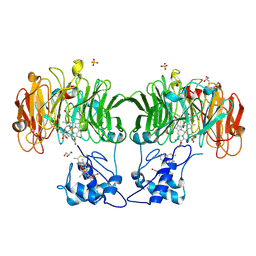 | | Cytochrome cd1 Nitrite Reductase, dioxygen complex | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ5
 
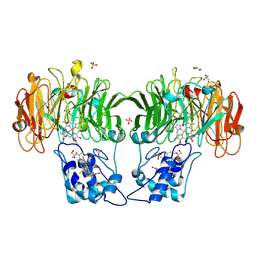 | | Cytochrome cd1 Nitrite Reductase, reoxidised enzyme | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
7F5J
 
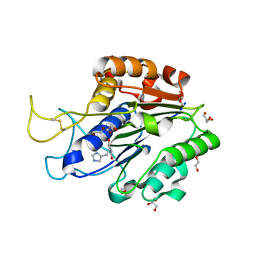 | | The crystal structure of VyPAL2-I244V, a more efficient mutant of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5Q
 
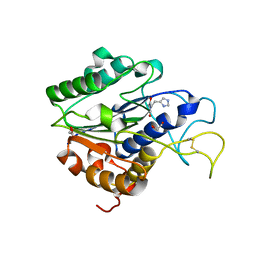 | | The crystal structure of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
4F7J
 
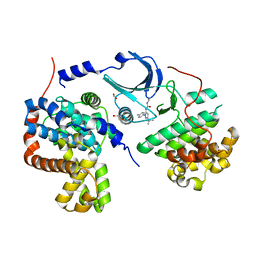 | | Crystal structure of human CDK8/CYCC in complex with compound 3 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(2-hydroxyethyl)urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(2-hydroxyethyl)urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1GQ1
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
1YLR
 
 | | The structure of E.coli nitroreductase with bound acetate, crystal form 1 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
4F6U
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 5 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
