1ECP
 
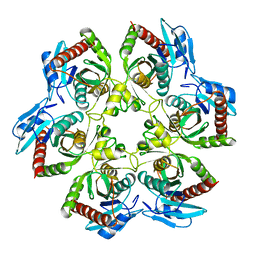 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-13 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.
Structure, 5, 1997
|
|
1SBE
 
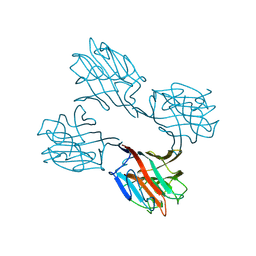 | | SOYBEAN AGGLUTININ FROM GLYCINE MAX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Dessen, A, Olsen, L.R, Gupta, D, Sabesan, S, Sacchettini, J.C, Brewer, C.F. | | Deposit date: | 1995-07-13 | | Release date: | 1998-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin.
Biochemistry, 36, 1997
|
|
1SBD
 
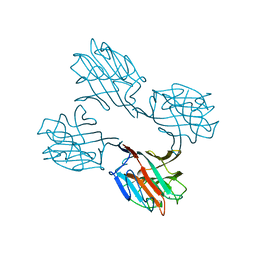 | | SOYBEAN AGGLUTININ COMPLEXED WITH 2,4-PENTASACCHARIDE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SOYBEAN AGGLUTININ, ... | | Authors: | Dessen, A, Olsen, L.R, Gupta, D, Sabesan, S, Brewer, C.F, Sacchettini, J.C. | | Deposit date: | 1995-07-13 | | Release date: | 1998-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin.
Biochemistry, 36, 1997
|
|
1PKM
 
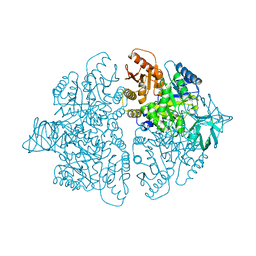 | |
1BPL
 
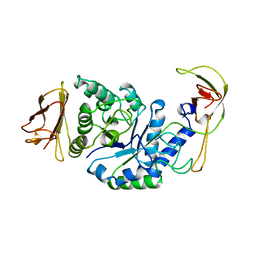 | | GLYCOSYLTRANSFERASE | | Descriptor: | ALPHA-1,4-GLUCAN-4-GLUCANOHYDROLASE | | Authors: | Machius, M, Wiegand, G, Huber, R. | | Deposit date: | 1995-07-13 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of calcium-depleted Bacillus licheniformis alpha-amylase at 2.2 A resolution.
J.Mol.Biol., 246, 1995
|
|
1OXA
 
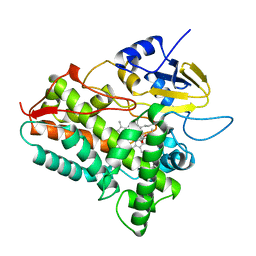 | | CYTOCHROME P450 (DONOR:O2 OXIDOREDUCTASE) | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, CYTOCHROME P450 ERYF, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 1995-07-14 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome P450eryF involved in erythromycin biosynthesis.
Nat.Struct.Biol., 2, 1995
|
|
1CPJ
 
 | |
1MML
 
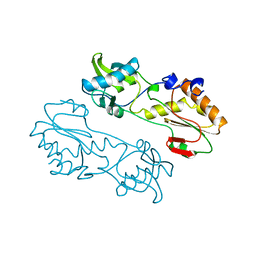 | | MECHANISTIC IMPLICATIONS FROM THE STRUCTURE OF A CATALYTIC FRAGMENT OF MMLV REVERSE TRANSCRIPTASE | | Descriptor: | MMLV REVERSE TRANSCRIPTASE | | Authors: | Georgiadis, M.M, Jessen, S.M, Ogata, C.M, Telesnitsky, A, Goff, S.P, Hendrickson, W.A. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase.
Structure, 3, 1995
|
|
1AJC
 
 | |
1BMG
 
 | |
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
1PFM
 
 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFN
 
 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1GCB
 
 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
217D
 
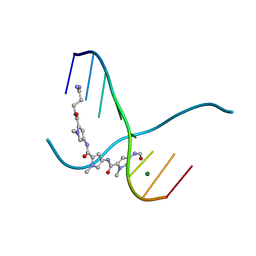 | |
216D
 
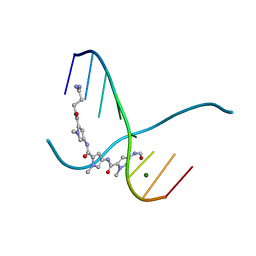 | |
218D
 
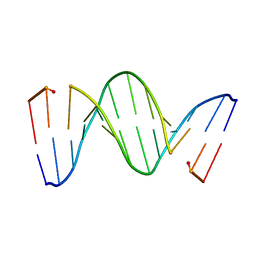 | | THE STRUCTURE OF A NEW CRYSTAL FORM OF A DNA DODECAMER CONTAINING T.(O6ME)G BASE PAIRS | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*AP*AP*TP*TP*CP*(6OG)P*CP*G)-3') | | Authors: | Vojtechovsky, J, Eaton, M.D, Gaffney, B, Jones, R, Berman, H.M. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a new crystal form of a DNA dodecamer containing T.(O6Me)G base pairs.
Biochemistry, 34, 1995
|
|
1MCY
 
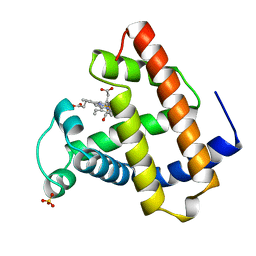 | |
1PFX
 
 | | PORCINE FACTOR IXA | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, FACTOR IXA | | Authors: | Brandstetter, H, Bauer, M, Huber, R, Lollar, P, Bode, W. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1CMF
 
 | | NMR SOLUTION STRUCTURE OF APO CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Finn, B.E, Evenas, J, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CMG
 
 | | NMR SOLUTION STRUCTURE OF CALCIUM-LOADED CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Evenas, J, Finn, B.E, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1BRE
 
 | | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN | | Descriptor: | BENCE-JONES KAPPA I PROTEIN BRE | | Authors: | Schormann, N, Benson, M.D. | | Deposit date: | 1995-07-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1GEN
 
 | | C-TERMINAL DOMAIN OF GELATINASE A | | Descriptor: | CALCIUM ION, CHLORIDE ION, GELATINASE A, ... | | Authors: | Libson, A.M, Gittis, A.G, Collier, I.E, Marmer, B.L, Goldberg, G.G, Lattman, E.E. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Nat.Struct.Biol., 2, 1995
|
|
226D
 
 | |
1SE2
 
 | |
