6YW7
 
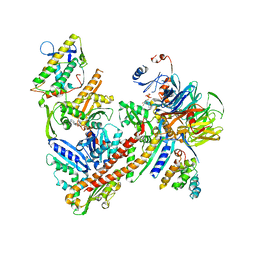 | | Cryo-EM structure of the ARP2/3 1A5C isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
3FM7
 
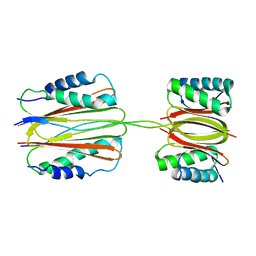 | | Quaternary Structure of Drosophila melanogaster IC/Tctex-1/LC8; Allosteric Interactions of Dynein Light Chains with Dynein Intermediate Chain | | Descriptor: | Dynein intermediate chain, cytosolic, Dynein light chain 1, ... | | Authors: | Hall, J.D, Karplus, P.A, Barbar, E.J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multivalency in the assembly of intrinsically disordered Dynein intermediate chain.
J.Biol.Chem., 284, 2009
|
|
3P9Y
 
 | |
3FRL
 
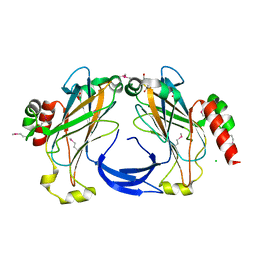 | | The 2.25 A crystal structure of LipL32, the major surface antigen of Leptospira interrogans serovar Copenhageni | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, CHLORIDE ION, LipL32, ... | | Authors: | Farah, C.S, Guzzo, C.R, Hauk, P, Ho, P.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp.
J.Mol.Biol., 390, 2009
|
|
7OAM
 
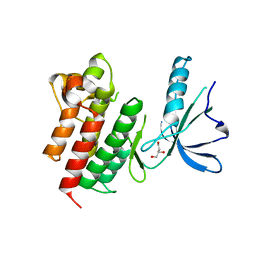 | | Kinase domain of MERTK in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(fluoranyl)phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schroeder, M, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
5FGO
 
 | |
7P37
 
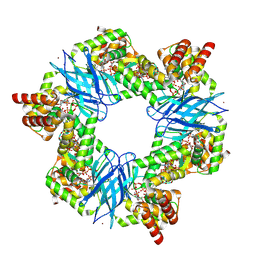 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3Q
 
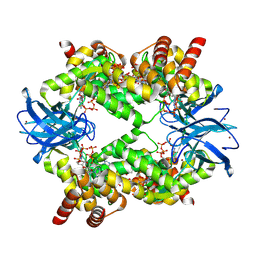 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3F
 
 | |
3GH6
 
 | |
6Z9U
 
 | | Crystal structure of a TSEN15-34 heterodimer. | | Descriptor: | GLYCEROL, tRNA-splicing endonuclease subunit Sen15, tRNA-splicing endonuclease subunit Sen34 | | Authors: | Trowitzsch, S, Sekulovski, S. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.10001779 Å) | | Cite: | Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia.
Nat Commun, 12, 2021
|
|
7OAJ
 
 | | Crystal structure of pseudokinase CASK in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(cyclopentylamino)-2-[(3,4-dichlorophenyl)methylamino]-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAI
 
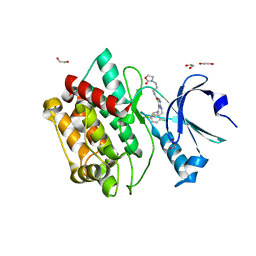 | | Crystal structure of pseudokinase CASK in complex with PFE-PKIS12 | | Descriptor: | 1,2-ETHANEDIOL, 4-(Cyclopentylamino)-2-[(2,5-dichlorophenyl)methylamino]-N-[3-(2-oxo-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAK
 
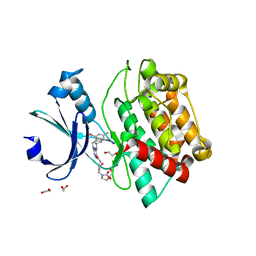 | | Crystal structure of pseudokinase CASK in complex with compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAL
 
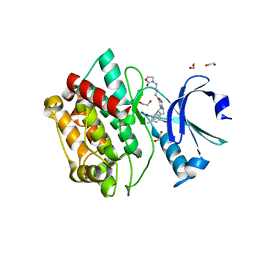 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
4YGX
 
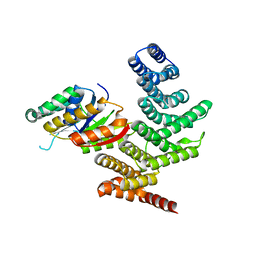 | |
7PA5
 
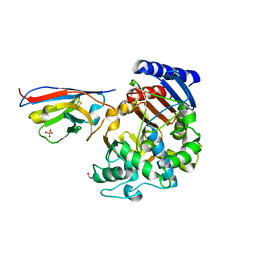 | | Complex between the beta-lactamase CMY-2 with an inhibitory nanobody | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Frederic Cawez, F.C, Frederic Kerff, F.K, Moreno Galleni, M.G, Raphael Herman, R.H. | | Deposit date: | 2021-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.184 Å) | | Cite: | Development of Nanobodies as Theranostic Agents against CMY-2-Like Class C beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
6Y79
 
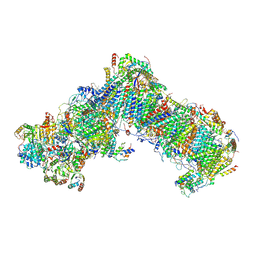 | | Cryo-EM structure of a respiratory complex I F89A mutant | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Essential role of accessory subunit LYRM6 in the mechanism of mitochondrial complex I.
Nat Commun, 11, 2020
|
|
6YD0
 
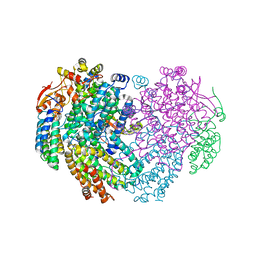 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, diferric state | | Descriptor: | FE (III) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
6YFB
 
 | | Virus-like particle of bacteriophage AVE016 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
3FYQ
 
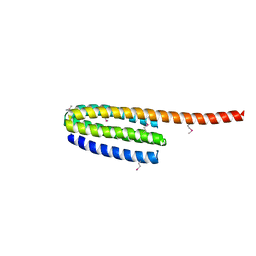 | | Structure of Drosophila melanogaster talin IBS2 domain (residues 1981-2168) | | Descriptor: | CG6831-PA (Talin) | | Authors: | Cheung, T.Y.S, Fairchild, M.J, Zarivach, R, Tanentzapf, G, Van Petegem, F. | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the talin integrin binding domain 2.
J.Mol.Biol., 387, 2009
|
|
7OSR
 
 | |
7OSW
 
 | |
6YFF
 
 | | Virus-like particle of Beihai levi-like virus 21 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFK
 
 | | Virus-like particle of bacteriophage ESE007 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
