6R4N
 
 | | Crystal structure of S. cerevisia Niemann-Pick type C protein NPC2 with ergosterol bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, ... | | Authors: | Winkler, M.B.L, Kidmose, R.T, Pedersen, B.P. | | Deposit date: | 2019-03-22 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight into Eukaryotic Sterol Transport through Niemann-Pick Type C Proteins.
Cell, 179, 2019
|
|
8C80
 
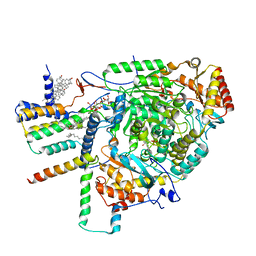 | | Cryo-EM structure of the yeast SPT-Orm1-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C81
 
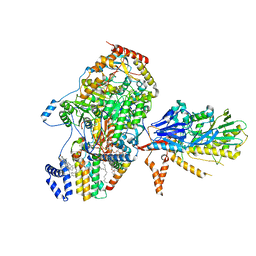 | | Cryo-EM structure of the yeast SPT-Orm1-Sac1 complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
9F40
 
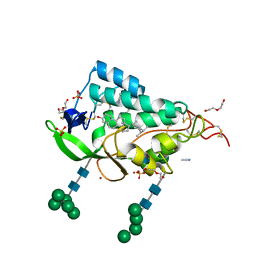 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with ergosterol bound | | Descriptor: | ACETONITRILE, DI(HYDROXYETHYL)ETHER, ERGOSTEROL, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C 1-related protein
To Be Published
|
|
6VGD
 
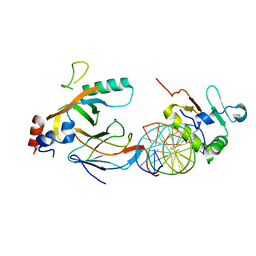 | |
6VG2
 
 | |
6VG8
 
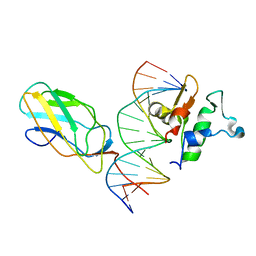 | |
6QV2
 
 | | Structure of ATPgS-bound outward-facing TM287/288 in complex with nanobody Nb_TM#2 | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (4.23 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
6QV1
 
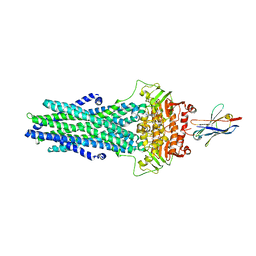 | | Structure of ATPgS-bound outward-facing TM287/288 in complex with nanobody Nb_TM1 | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
6QUZ
 
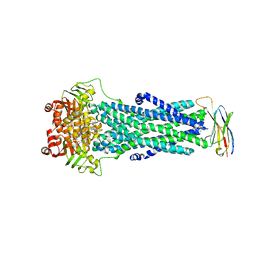 | | Structure of ATPgS-bound outward-facing TM287/288 in complex with sybody Sb_TM35 | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
6QV0
 
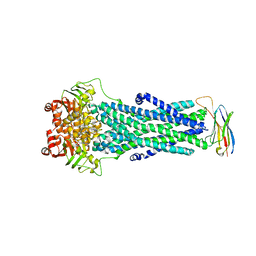 | | Structure of ATP-bound outward-facing TM287/288 in complex with sybody Sb_TM35 | | Descriptor: | ABC transporter, ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
4X8M
 
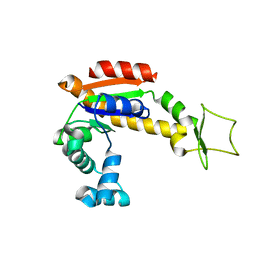 | | Crystal structure of E. coli Adenylate kinase Y171W mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8O
 
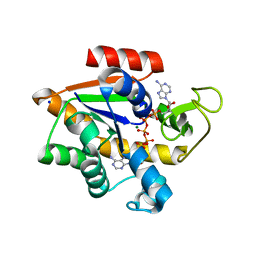 | | Crystal structure of E. coli Adenylate kinase Y171W mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8L
 
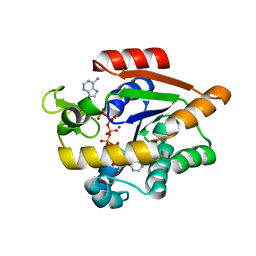 | | Crystal structure of E. coli Adenylate kinase P177A mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8H
 
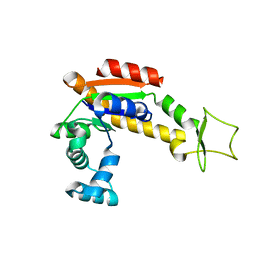 | | Crystal structure of E. coli Adenylate kinase P177A mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4A7D
 
 | | X-ray crystal structure of HEWL flash-cooled at high pressure | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Burkhardt, A, Warmer, M, Panneerselvam, S, Wagner, A, Reimer, R, Hohenberg, H, Meents, A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Fast High-Pressure Freezing of Protein Crystals in Their Mother Liquor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4UUN
 
 | |
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTB
 
 | | 2-propylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-propylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTF
 
 | | 4-chloro-1h-pyrazole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4-chloro-1H-pyrazole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTD
 
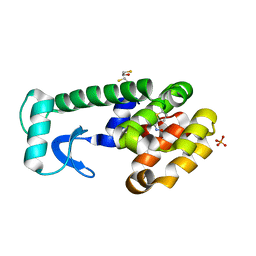 | | (Z)-Thiophene-2-carboxaldoxime in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (NZ)-N-(thiophen-2-ylmethylidene)hydroxylamine, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HU8
 
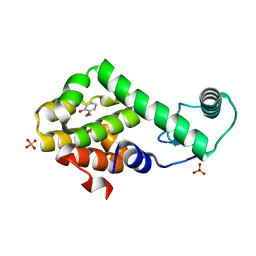 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT9
 
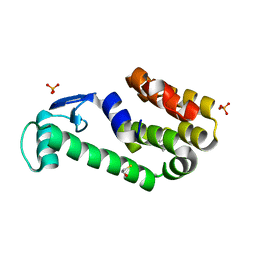 | | 2-methoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Guaiacol, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
