5F63
 
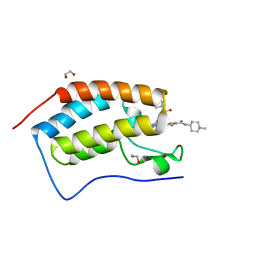 | | Crystal structure of the first bromodomain of human BRD4 in complex with SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
1MRQ
 
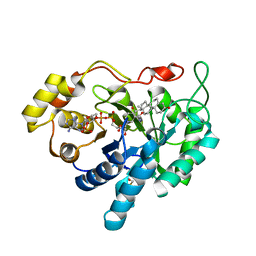 | | Crystal structure of human 20alpha-HSD in ternary complex with NADP and 20alpha-hydroxy-progesterone | | Descriptor: | Aldo-keto reductase family 1 member C1, BETA-MERCAPTOETHANOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Couture, J.F, Legrand, P, Cantin, L, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human 20alpha-hydroxysteroid dehydrogenase: crystallographic and site-directed mutagenesis studies lead to the identification of an alternative binding site for C21-steroids.
J.Mol.Biol., 331, 2003
|
|
6DWN
 
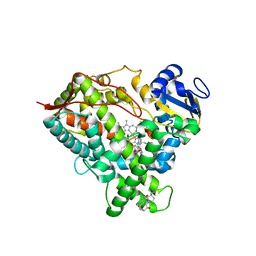 | | Structure of Human Cytochrome P450 1A1 with Erlotinib | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Cytochrome P450 1A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands.
J. Biol. Chem., 293, 2018
|
|
3JYP
 
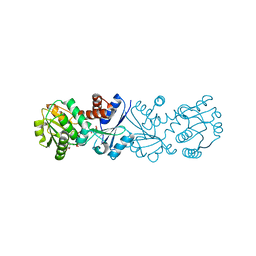 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
6FKS
 
 | | Crystal structure of a dye-decolorizing peroxidase from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.60000467 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
1T6M
 
 | | X-ray Structure of the R70D PI-PLC enzyme: Insight into the role of calcium and surrounding amino acids on active site geometry and catalysis. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, CALCIUM ION | | Authors: | Apiyo, D, Zhao, L, Tsai, M.-D, Selby, T.L. | | Deposit date: | 2004-05-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | X-ray Structure of the R69D Phosphatidylinositol-Specific Phospholipase C Enzyme: Insight into the Role of Calcium and Surrounding Amino Acids in Active Site Geometry and Catalysis.
Biochemistry, 44, 2005
|
|
6C0T
 
 | | Crystal structure of cGMP-dependent protein kinase Ialpha (PKG Ialpha) catalytic domain bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, TRIETHYLENE GLYCOL, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
5HG1
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
1HMT
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, STEARIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
3UK2
 
 | |
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
1GD4
 
 | | SOLUTION STRUCTURE OF P25S CYSTATIN A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
9DM1
 
 | | Mycobacterial supercomplex malate:quinone oxidoreductase assembly | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Di Trani, J.M, Rubinstein, J.L. | | Deposit date: | 2024-09-11 | | Release date: | 2024-10-23 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of native membranes reveals an intimate connection between the Krebs cycle and aerobic respiration in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1LDE
 
 | |
6S5V
 
 | | Crystal structure of the Cap-Midlink region of the H5N1 Influenza A virus polymerase in complex with a Cap-domain binding analogue | | Descriptor: | (1~{S},2~{S},3~{S},6~{R})-2-[[2-[5,7-bis(fluoranyl)-1~{H}-indol-3-yl]-5-fluoranyl-pyrimidin-4-yl]amino]-3,6-dimethyl-cyclohexane-1-carboxylic acid, GLYCEROL, POTASSIUM ION, ... | | Authors: | Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-07-02 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Novel Indoles Targeting the Influenza PB2 Cap Binding Region.
J.Med.Chem., 62, 2019
|
|
7DAY
 
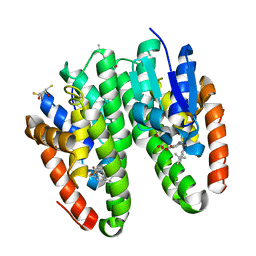 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-, and GSH-bound form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
6BVK
 
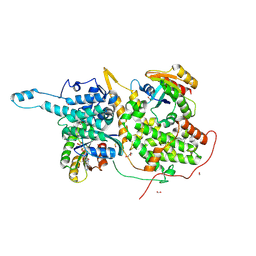 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | FORMIC ACID, GLYCEROL, GTPase HRas, ... | | Authors: | Phan, J, Abbott, J, Fesik, S.W. | | Deposit date: | 2017-12-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Discovery of Aminopiperidine Indoles That Activate the Guanine Nucleotide Exchange Factor SOS1 and Modulate RAS Signaling.
J. Med. Chem., 61, 2018
|
|
7EJ2
 
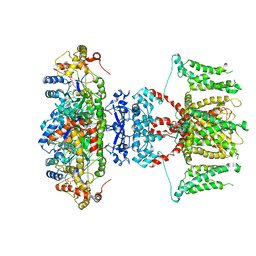 | | human voltage-gated potassium channel KV1.3 H451N mutant | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
6UNG
 
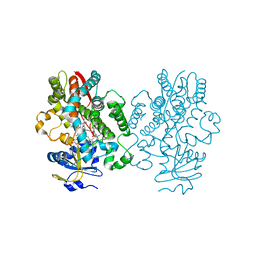 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(naphthalen-1-yl)-3-{[(2R)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
1AMY
 
 | |
5UCQ
 
 | | The structure of archaeal Inorganic Pyrophosphatase in complex with pyrophosphate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Inoguchi, N, Hughes, R.C, Meehan, E.J, Ng, J.D. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of archaeal Inorganic Pyrophosphatase in complex with pyrophosphate
To Be Published
|
|
5UGU
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT506 | | Descriptor: | 2-[4-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5I05
 
 | |
5YTU
 
 | |
5YSR
 
 | | Ethanolamine ammonia-lyase, AdoCbl/2-amino-1-propanol | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Ethanolamine ammonia-lyase heavy chain, ... | | Authors: | Shibata, N. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct Participation of a Peripheral Side Chain of a Corrin Ring in Coenzyme B12Catalysis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
