6N1V
 
 | |
6MP1
 
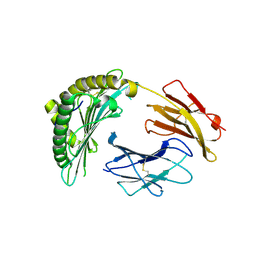 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with the mutant TRP1-K8 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TRP1-K8 peptide, Beta-2-microglobulin,H-2 class I histocompatibility antigen, ... | | Authors: | Clancy-Thompson, E, Devlin, C.A, Birnbaum, M.E, Dougan, S.K. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Altered Binding of Tumor Antigenic Peptides to MHC Class I Affects CD8+T Cell-Effector Responses.
Cancer Immunol Res, 6, 2018
|
|
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
8OH7
 
 | |
6N8X
 
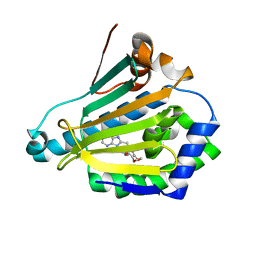 | | Hsp90-alpha bound to PU-11-trans | | Descriptor: | 9-[(2E)-but-2-en-1-yl]-8-[(3,4,5-trimethoxyphenyl)methyl]-9H-purin-6-amine, Heat shock protein HSP 90-alpha | | Authors: | Huck, J.D, Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2018-11-30 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49484479 Å) | | Cite: | Structures of Hsp90 alpha and Hsp90 beta bound to a purine-scaffold inhibitor reveal an exploitable residue for drug selectivity.
Proteins, 87, 2019
|
|
5D76
 
 | |
6MUL
 
 | | Murine PI3K delta kinsae domain - cpd 1 | | Descriptor: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
6MPW
 
 | |
6MQG
 
 | |
6N0D
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluorobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575
To Be Published
|
|
5DFN
 
 | | Structure of Tetrahymena Telomerase P45 C-terminal domain | | Descriptor: | Telomerase associated protein p45 | | Authors: | Chan, H, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions.
Science, 350, 2015
|
|
5DDO
 
 | |
5DE5
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA. | | Descriptor: | Fragile X mental retardation protein 1, POTASSIUM ION, sc1 | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.0011 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CV1
 
 | | C. elegans PGL-1 Dimerization Domain | | Descriptor: | P granule abnormality protein 1 | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CT3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with 2FA | | Descriptor: | 3-hydroxypropane-1-sulfonic acid, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5D6I
 
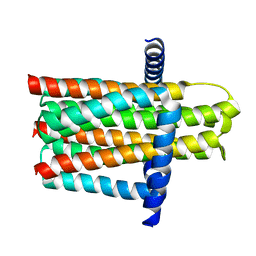 | | DgkA - CIM | | Descriptor: | Diacylglycerol kinase | | Authors: | Ma, P, Caffrey, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5CS3
 
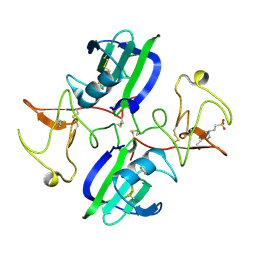 | | The structure of the NK1 fragment of HGF/SF complexed with (H)EPPS | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5D9Q
 
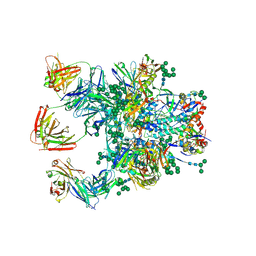 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 and scFv NIH45-46 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Ward, A.B, Wilson, I.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
5D1Q
 
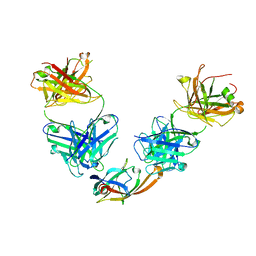 | | IsdB NEAT2 bound by clone D2-06 | | Descriptor: | D2-06 Heavy Chain, D2-06 Light Chain, Iron-regulated surface determinant protein B, ... | | Authors: | Deng, X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-08-10 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire.
Nat Commun, 7, 2016
|
|
6NE7
 
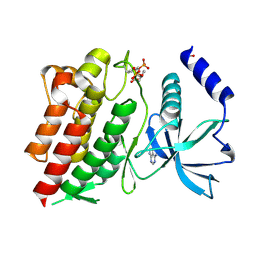 | | Structure of G810A mutant of RET protein tyrosine kinase domain. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
8OG0
 
 | |
6NPK
 
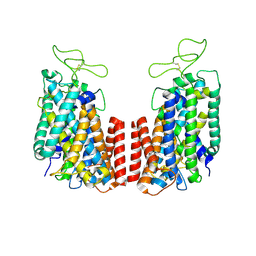 | | Structure of the TM domain | | Descriptor: | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2019-08-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6NQ1
 
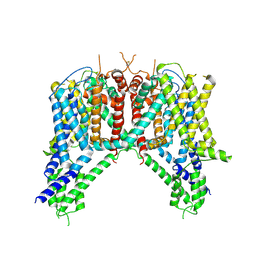 | | Cryo-EM structure of human TPC2 channel in the apo state | | Descriptor: | Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
7QQJ
 
 | | Sucrose phosphorylase from Jeotgalibaca ciconiae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose phosphorylase | | Authors: | Ubiparip, Z, Capra, N, Rozeboom, H.J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sucrose phosphorylase from Jeotgalibaca ciconiae
To Be Published
|
|
