2GAZ
 
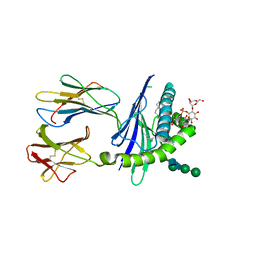 | | Mycobacterial lipoglycan presentation by CD1d | | Descriptor: | (2R)-3-[(HYDROXY{[(2R,3R,5S,6R)-3,4,5-TRIHYDROXY-2,6-BIS(ALPHA-D-MANNOPYRANOSYLOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]PROPAN E-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d.
J.Immunol., 177, 2006
|
|
2GB0
 
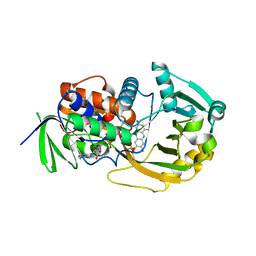 | | Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Trickey, P, Wagner, M.A, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric Sarcosine Oxidase: Structure of a Covalently Flavinylated Amine Oxidizing Enzyme
Structure, 7, 1999
|
|
2GB1
 
 | |
2GB2
 
 | | The P52G mutant of amicyanin in the Cu(II) state. | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-03-09 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
2GB3
 
 | |
2GB4
 
 | |
2GB5
 
 | |
2GB7
 
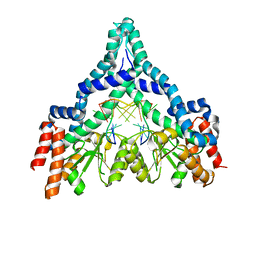 | | Metal-depleted Ecl18kI in complex with uncleaved, modified DNA | | Descriptor: | DNA STRAND 1, DNA STRAND 2, R.Ecl18kI | | Authors: | Bochtler, M, Szczepanowski, R.H, Tamulaitis, G, Grazulis, S, Czapinska, H, Manakova, E, Siksnys, V. | | Deposit date: | 2006-03-10 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease
Embo J., 25, 2006
|
|
2GB8
 
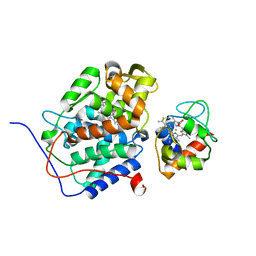 | | Solution structure of the complex between yeast iso-1-cytochrome c and yeast cytochrome c peroxidase | | Descriptor: | Cytochrome c iso-1, Cytochrome c peroxidase, HEME C, ... | | Authors: | Volkov, A.N, Worrall, J.A.R, Ubbink, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GB9
 
 | | d(CGTACG)2 crosslinked bis-acridine complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9,9'-(HEXANE-1,6-DIYLDIIMINO)BIS{N-[2-(DIMETHYLAMINO)ETHYL]ACRIDINE-4-CARBOXAMIDE}, STRONTIUM ION | | Authors: | Hopcroft, N.H, Brogden, A.L, Searcey, M, Cardin, C.J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystallographic study of DNA duplex cross-linking: simultaneous binding to two d(CGTACG)2 molecules by a bis(9-aminoacridine-4-carboxamide) derivative.
Nucleic Acids Res., 34, 2006
|
|
2GBA
 
 | | Reduced Cu(I) form at pH 4 of P52G mutant of amicyanin | | Descriptor: | COPPER (I) ION, amicyanin | | Authors: | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-03-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
2GBB
 
 | | Crystal structure of secreted chorismate mutase from Yersinia pestis | | Descriptor: | CITRIC ACID, SULFATE ION, putative chorismate mutase | | Authors: | Ladner, J.E, Reddy, P.T, Nelson, B.C, Robinson, H, Kim, S.-K. | | Deposit date: | 2006-03-10 | | Release date: | 2007-04-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H(37)R(v) and the secreted chorismate mutase (y2828) from Yersinia pestis.
Febs J., 275, 2008
|
|
2GBC
 
 | | Native DPP-IV (CD26) from Rat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
2GBF
 
 | | rat dpp-IV with alkynyl cyanopyrrolidine #1 | | Descriptor: | (1S)-2-[(2S,5R)-2-(AMINOMETHYL)-5-ETHYNYLPYRROLIDIN-1-YL]-1-CYCLOPENTYL-2-OXOETHANAMINE, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
2GBG
 
 | | rat DPP-IV with alkynyl cyanopyrrolidine #2 | | Descriptor: | (1S)-2-[(2S,5R)-2-(AMINOMETHYL)-5-PROP-1-YN-1-YLPYRROLIDIN-1-YL]-1-CYCLOPENTYL-2-OXOETHANAMINE, Dipeptidyl peptidase 4, SULFATE ION | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
2GBI
 
 | | rat DPP-IV with xanthine inhibitor 4 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
2GBJ
 
 | |
2GBK
 
 | |
2GBL
 
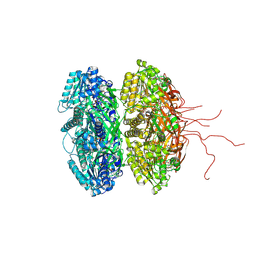 | | Crystal Structure of Full Length Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.R, Pattanayek, S, Xu, Y, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2006-03-10 | | Release date: | 2007-01-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods.
Embo J., 25, 2006
|
|
2GBM
 
 | | Crystal Structure of the 35-36 8 Glycine Insertion Mutant of Ubiquitin | | Descriptor: | ARSENIC, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
2GBN
 
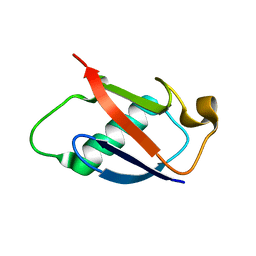 | |
2GBO
 
 | |
2GBP
 
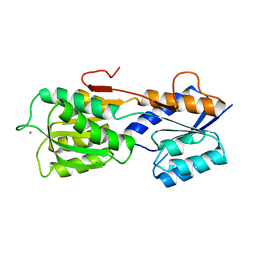 | |
2GBQ
 
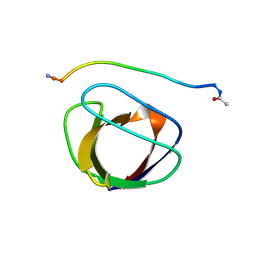 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
