6NRE
 
 | | Monomeric Lipocalin Can F 6 | | Descriptor: | Lipocalin-Can f 6 allergen | | Authors: | Clayton G, M, Kappler J, W, Chan, S. | | Deposit date: | 2019-01-23 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural characteristics of lipocalin allergens: Crystal structure of the immunogenic dog allergen Can f 6.
Plos One, 14, 2019
|
|
1BBP
 
 | | MOLECULAR STRUCTURE OF THE BILIN BINDING PROTEIN (BBP) FROM PIERIS BRASSICAE AFTER REFINEMENT AT 2.0 ANGSTROMS RESOLUTION. | | Descriptor: | BILIN BINDING PROTEIN, BILIVERDIN IX GAMMA CHROMOPHORE | | Authors: | Huber, R, Schneider, M, Mayr, I, Mueller, R, Deutzmann, R, Suter, F, Zuber, H, Falk, H, Kayser, H. | | Deposit date: | 1990-09-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution.
J.Mol.Biol., 198, 1987
|
|
6NKQ
 
 | |
1B8E
 
 | |
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
6NNX
 
 | |
6NNY
 
 | |
1B56
 
 | | HUMAN RECOMBINANT EPIDERMAL FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Van Tilbeurgh, H, Hohoff, C, Borchers, T, Spener, F. | | Deposit date: | 1999-01-12 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein.
Biochemistry, 38, 1999
|
|
1AQB
 
 | | RETINOL-BINDING PROTEIN (RBP) FROM PIG PLASMA | | Descriptor: | CADMIUM ION, RETINOL, RETINOL-BINDING PROTEIN | | Authors: | Zanotti, G, Panzalorto, M, Marcato, A, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pig plasma retinol-binding protein at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1B0O
 
 | |
1BEB
 
 | |
6ON5
 
 | |
6ON8
 
 | |
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
1A18
 
 | | PHENANTHROLINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-23 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
6NOE
 
 | |
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
1BSO
 
 | | 12-BROMODODECANOIC ACID BINDS INSIDE THE CALYX OF BOVINE BETA-LACTOGLOBULIN | | Descriptor: | 12-BROMODODECANOIC ACID, PROTEIN (BOVINE BETA-LACTOGLOBULIN A) | | Authors: | Qin, B.Y, Creamer, L.K, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | 12-Bromododecanoic acid binds inside the calyx of bovine beta-lactoglobulin.
FEBS Lett., 438, 1998
|
|
1BSY
 
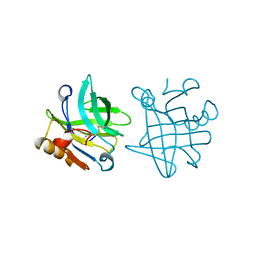 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 7.1 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
6ON7
 
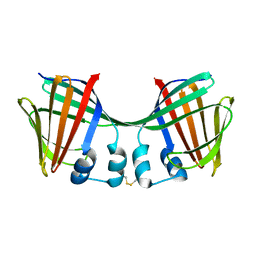 | |
2OVD
 
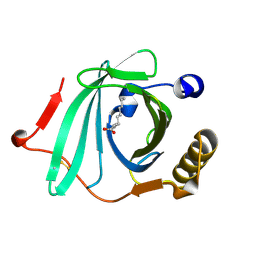 | | Crystal Structure of Human Complement Protein C8gamma with Laurate | | Descriptor: | Complement component 8, gamma polypeptide, LAURIC ACID | | Authors: | Chiswell, B, Lovelace, L.L, Brannen, C, Ortlund, E.A, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural features of the ligand binding site on human complement protein C8gamma: A member of the lipocalin family
Biochim.Biophys.Acta, 1774, 2007
|
|
2OVE
 
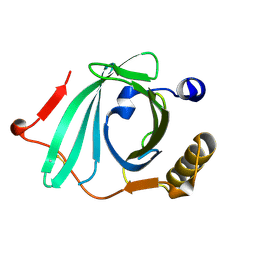 | | Crystal Structure of Recombinant Human Complement Protein C8gamma | | Descriptor: | Complement component 8, gamma polypeptide | | Authors: | Chiswell, B, Lovelace, L.L, Brannen, C, Ortlund, E.A, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural features of the ligand binding site on human complement protein C8gamma: A member of the lipocalin family
Biochim.Biophys.Acta, 1774, 2007
|
|
2OVA
 
 | | X-ray structure of Human Complement Protein C8gamma Y83W Mutant | | Descriptor: | Complement component 8, gamma polypeptide | | Authors: | Chiswell, B, Lovelace, L.L, Brannen, C, Ortlund, E.A, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural features of the ligand binding site on human complement protein C8gamma: A member of the lipocalin family
Biochim.Biophys.Acta, 1774, 2007
|
|
2NNQ
 
 | |
2NNE
 
 | | The Structural Identification of the Interaction Site and Functional State of RBP for its Membrane Receptor | | Descriptor: | CADMIUM ION, GLYCEROL, Major urinary protein 2 | | Authors: | Redondo, C, Bingham, R.J, Vouropoulou, M, Homans, S.W, Findlay, J.B. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the retinol-binding protein (RBP) interaction site and functional state of RBPs for the membrane receptor.
Faseb J., 22, 2008
|
|
