7JTT
 
 | |
7JMQ
 
 | | The external aldimine form of the mutant beta-S377A Salmonella thypi tryptophan synthase in open conformation showing dual side chain conformations for the residue beta-Q114, sodium ion at the metal coordination site, and F9 inhibitor at the alpha-site. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The external aldimine form of mutant beta-S377A Salmonella thypi tryptophan synthase in open conformation showing dual side chain conformations for the residue beta-Q114, sodium ion at the metal coordination site, and F9 inhibitor at the alpha-site. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
3IAP
 
 | | E. coli (lacZ) beta-galactosidase (E416Q) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
7DPS
 
 | |
3IAQ
 
 | | E. coli (lacz) beta-galactosidase (E416V) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
3T0D
 
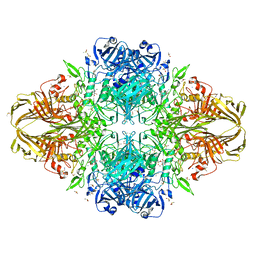 | | E.coli (lacZ) beta-galactosidase (S796T) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ser-796 of Beta-galactosidase (E. coli) plays a Key role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2Q
 
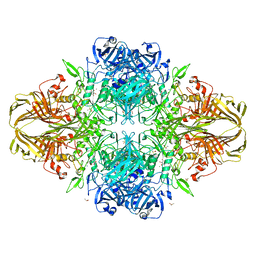 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2P
 
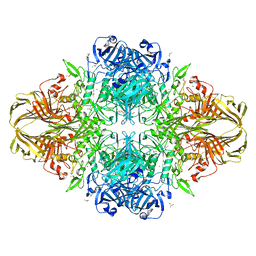 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0B
 
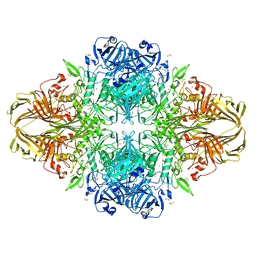 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2O
 
 | | E. coli (lacZ) beta-galactosidase (S796D) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0A
 
 | | E. coli (LacZ) beta-galactosidase (S796T) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T09
 
 | | E. coli (LacZ) beta-galactosidase (S796A) galactonolactone complex | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
1LCZ
 
 | | streptavidin-BCAP complex | | Descriptor: | E-AMINO BIOTINYL CAPROIC ACID, Streptavidin | | Authors: | Livnah, O, Pazy, Y, Bayer, E.A, Wilchek, M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
2YIR
 
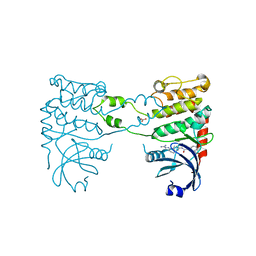 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1352 | | Descriptor: | (E)-N-(5-(2-CARBAMIMIDOYLHYDRAZONO)-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
3T08
 
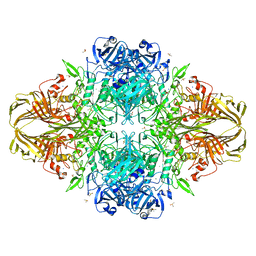 | | E. coli (LacZ) beta-galactosidase (S796A) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
1YA4
 
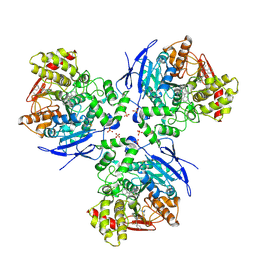 | | Crystal Structure of Human Liver Carboxylesterase 1 in complex with tamoxifen | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CES1 protein, ... | | Authors: | Fleming, C.D, Bencharit, S, Edwards, C.C, Hyatt, J.L, Morton, C.L, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2004-12-17 | | Release date: | 2005-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into drug processing by human carboxylesterase 1: tamoxifen, mevastatin, and inhibition by benzil.
J.Mol.Biol., 352, 2005
|
|
3GK2
 
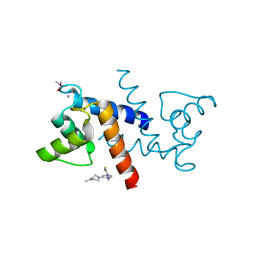 | | X-ray structure of bovine SBi279,Ca(2+)-S100B | | Descriptor: | (Z)-2-[2-(4-methylpiperazin-1-yl)benzyl]diazenecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
4PKK
 
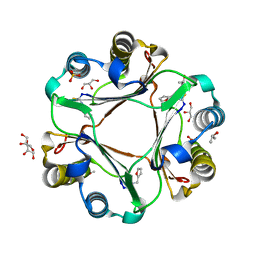 | |
4AVG
 
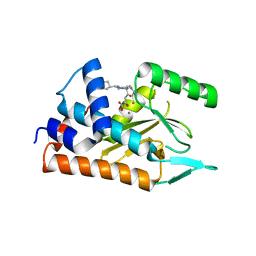 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with diketo compound 2 | | Descriptor: | (Z)-4-[4-[(4-chlorophenyl)methyl]-1-(cyclohexylmethyl)piperidin-4-yl]-2-oxidanyl-4-oxidanylidene-but-2-enoic acid, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-05-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
2Q94
 
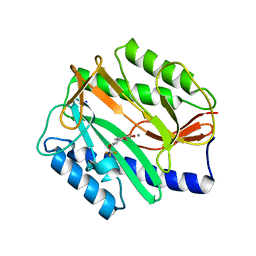 | | E. coli methionine aminopeptidase Mn-form with inhibitor A04 | | Descriptor: | 5-[2-(TRIFLUOROMETHOXY)PHENYL]-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
3ODI
 
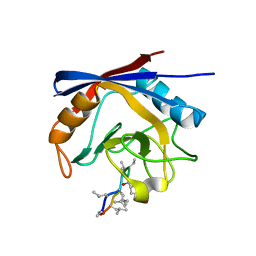 | | Crystal structure of cyclophilin A in complex with Voclosporin E-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Q95
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A05 | | Descriptor: | 5-(2-CHLORO-4-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q96
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A18 | | Descriptor: | 5-(2-CHLOROBENZYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q92
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B23 | | Descriptor: | 5-(2-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q93
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B21 | | Descriptor: | 5-(2-METHOXYPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
