6QWB
 
 | | Crystal structure of KPC-4 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2~{S})-5-azanylidene-2-(piperidin-4-ylcarbamoyl)piperidine-1-carboxylic acid, Beta-lactamase, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
7XDF
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor EA | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7V6Q
 
 | | Crystal structure of sNASP-ASF1A-H3.1-H4 complex | | Descriptor: | GLYCEROL, Histone H3.1, Histone H4, ... | | Authors: | Liu, C.P, Xu, R.M. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distinct histone H3-H4 binding modes of sNASP reveal the basis for cooperation and competition of histone chaperones.
Genes Dev., 35, 2021
|
|
5X1I
 
 | |
1LX6
 
 | |
6HKB
 
 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 3 | | Descriptor: | (1~{R},2~{S})-2-[methyl-[(~{R})-(2-methylpropan-2-yl)oxy-oxidanyl-methyl]amino]-2-phenyl-1-(2-sulfanylethylamino)ethanol, 14-3-3 protein sigma, Estrogen receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
4ZF4
 
 | |
6EPS
 
 | | The ATAD2 bromodomain in complex with compound UZH-DQ41 | | Descriptor: | (2~{R})-~{N}-[5-[3,5-bis(oxidanyl)phenyl]-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Hitting a moving target: simulation and crystallography study of ATAD2 bromodomain blockers
To Be Published
|
|
6QW8
 
 | | Crystal structure of CTX-M-15 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QRM
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKRRAA substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
9DY4
 
 | |
6R3P
 
 | | Crystal structure of human DMC1 ATPase domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Dunce, J.M, Davies, O.R. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BRCA2 stabilises RAD51 and DMC1 nucleoprotein filaments through a conserved interaction mode.
Nat Commun, 15, 2024
|
|
6HEI
 
 | |
8DLB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | Descriptor: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
6R9D
 
 | |
8DMD
 
 | |
5DXY
 
 | | Crystal structure of Dbr2 | | Descriptor: | Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE, [[(2R,3S,4R,5R)-5-(5-aminocarbonyl-3,4-dihydro-2H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
5MTP
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT514 | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
6HS1
 
 | | EthR2 in complex with compound 9 (BDM76060) | | Descriptor: | (1~{R},5~{S})-9-[2-(4-chlorophenyl)ethyl]-9-azabicyclo[3.3.1]nonan-3-one, Probable transcriptional regulatory protein | | Authors: | Wintjens, R, Wohlkonig, A, Tanina, A. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A fragment-based approach towards the discovery of N-substituted tropinones as inhibitors of Mycobacterium tuberculosis transcriptional regulator EthR2.
Eur J Med Chem, 167, 2019
|
|
7PH2
 
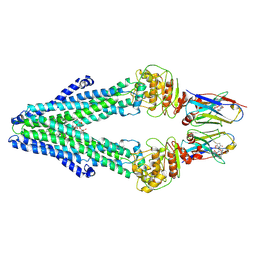 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position A60C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase, ... | | Authors: | Januliene, D, Parey, K, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PH7
 
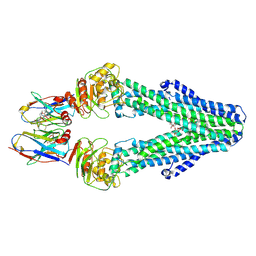 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position T68C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-binding transport protein multicopy suppressor of htrB, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
4QXM
 
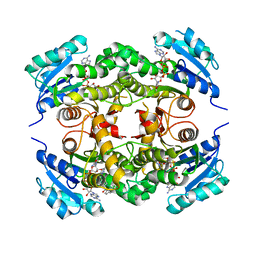 | | Crystal structure of the InhA:GSK_SB713 complex | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-(2-chloro-4-fluorobenzyl)-4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]benzamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gulten, G, Sacchettini, J.C. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | N-Benzyl-4-((heteroaryl)methyl)benzamides: A New Class of Direct NADH-Dependent 2-trans Enoyl-Acyl Carrier Protein Reductase (InhA) Inhibitors with Antitubercular Activity.
Chemmedchem, 11, 2016
|
|
6HYM
 
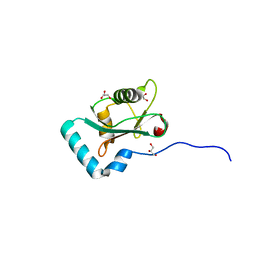 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
5MWH
 
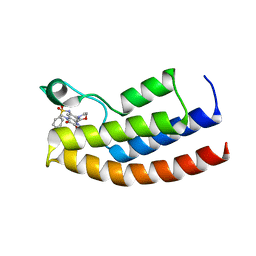 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ089 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-7-morpholin-4-yl-2,3-bis(oxidanylidene)quinoxalin-6-yl]-4-(2-methylpropyl)benzenesulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
6HIW
 
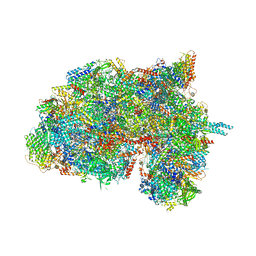 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete small mitoribosomal subunit in complex with mt-IF-3 | | Descriptor: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ramrath, D, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
