9MVY
 
 | | Crystal structure of ZMET2 in complex with unmethylated CTG DNA and a histone H3Kc9me2 peptide | | Descriptor: | C49 ssDNA, DNA (cytosine-5)-methyltransferase 1, Histone H3.2, ... | | Authors: | Herle, G, Fang, J, Song, J. | | Deposit date: | 2025-01-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an Unfavorable de Novo DNA Methylation Complex of Plant Methyltransferase ZMET2.
J.Mol.Biol., 437, 2025
|
|
6A0H
 
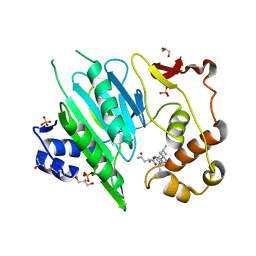 | |
5F68
 
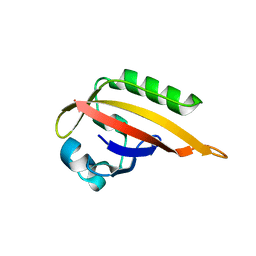 | |
7U4N
 
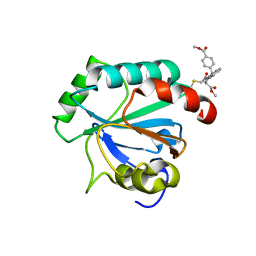 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
6GMQ
 
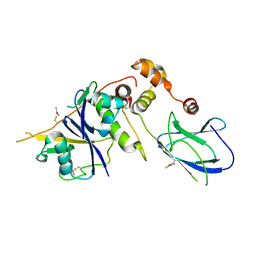 | | pVHL:EloB:EloC in complex with (4-(1H-pyrrol-1-yl)phenyl)methanol | | Descriptor: | (4-pyrrol-1-ylphenyl)methanol, ACETATE ION, Elongin-B, ... | | Authors: | Van Molle, I, Lucas, X, Ciulli, A. | | Deposit date: | 2018-05-28 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Surface Probing by Fragment-Based Screening and Computational Methods Identifies Ligandable Pockets on the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.
J. Med. Chem., 61, 2018
|
|
6GRL
 
 | |
7OR1
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
8U1W
 
 | | Structure of Norovirus (Hu/GII.4/Sydney/NSW0514/2012/AU) protease bound to inhibitor NV-004 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidase C37, ... | | Authors: | Eruera, A.R, Campbell, A.C, Krause, K.L. | | Deposit date: | 2023-09-03 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Inhibitor-Bound GII.4 Sydney 2012 Norovirus 3C-Like Protease.
Viruses, 15, 2023
|
|
6A0F
 
 | |
9GR0
 
 | | Interaction with AK2A links AIFM1 to cellular energy metabolism. The cryo-EM structure of dimeric AIFM1 bound by AK2A. | | Descriptor: | Adenylate kinase 2, mitochondrial, Apoptosis-inducing factor 1, ... | | Authors: | Rothemann, R.A, Pavlenko, E.A, Gerlich, S, Grobushkin, P, Mostert, S, Stobbe, D, Racho, J, Stillger, K, Lapacz, K, Petrungaro, C, Dengjel, J, Neundorf, I, Bano, D, Mondal, M, Weiss, K, Ehninger, D, Nguyen, T.H.D, Poepsel, S.P, Riemer, J. | | Deposit date: | 2024-09-10 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Interaction with AK2A links AIFM1 to cellular energy metabolism.
Mol.Cell, 85, 2025
|
|
7ZVE
 
 | | K403 acetylated glucose-6-phosphate dehydrogenase (G6PD) | | Descriptor: | COPPER (II) ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wu, F, Muskat, N.H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
6WYG
 
 | |
8YZZ
 
 | | The structure of HLA-A*2402 complex with peptide from SARS-CoV-2 S448-456 NYNYLYRLF(Prototype) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1 | | Authors: | Shang, B.L, Zhang, J.N, Tian, J.M, Liu, J. | | Deposit date: | 2024-04-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
5H5L
 
 | | Structure of prostaglandin synthase D of Nilaparvata lugens | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, S, Nakagawa, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular structure of a prostaglandin D synthase requiring glutathione from the brown planthopper, Nilaparvata lugens
Biochem. Biophys. Res. Commun., 492, 2017
|
|
6QJU
 
 | |
9LWF
 
 | | Cryo-EM structure of dual sensor bound GATOR2 complex | | Descriptor: | Cytosolic arginine sensor for mTORC1 subunit 1, GATOR2 complex protein MIOS, GATOR2 complex protein WDR24, ... | | Authors: | Su, M.-Y. | | Deposit date: | 2025-02-14 | | Release date: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structures of amino acid sensors bound to the human GATOR2 complex.
Cell Rep, 44, 2025
|
|
6QHK
 
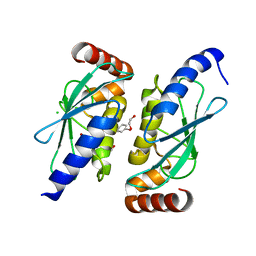 | |
5K4Z
 
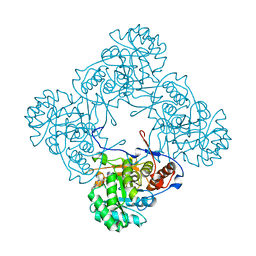 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
6HGG
 
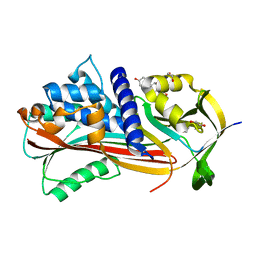 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-III: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
9GJY
 
 | |
8U4P
 
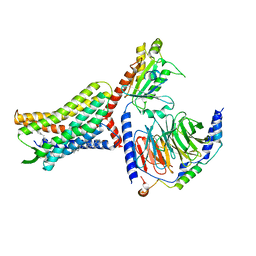 | | Structure of AMD3100-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6NMF
 
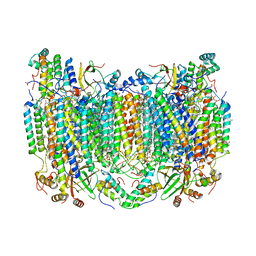 | | SFX structure of reduced cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-10 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HGF
 
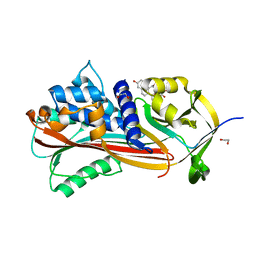 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-II: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6A0E
 
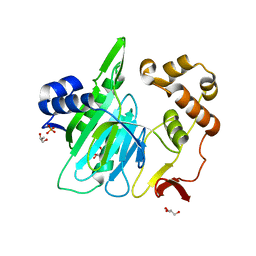 | |
6NMP
 
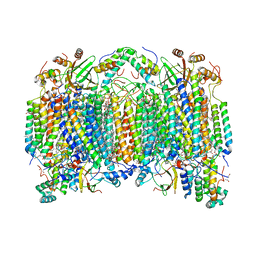 | | SFX structure of oxidized cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
