7WWD
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with squalene | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WVT
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1GLP
 
 | |
6T46
 
 | | Structure of the Rap conjugation gene regulator of the plasmid pLS20 in complex with the Phr* peptide | | Descriptor: | CHLORIDE ION, Quorum-sensing secretion protein (processed), Response regulator aspartate phosphatase, ... | | Authors: | Crespo, I, Bernardo, N, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2019-10-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization.
Nucleic Acids Res., 48, 2020
|
|
7MN0
 
 | |
7MKC
 
 | |
4ZK0
 
 | | Psoriasis pathogenesis - Pso p27 constitute a compact structure forming large aggregates. High pH structure | | Descriptor: | Serpin B4, ZINC ION | | Authors: | Helland, R, Lysvand, H, Slupphaug, G, Iversen, O.J. | | Deposit date: | 2015-04-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Psoriasis pathogenesis - Pso p27 constitutes a compact structure forming large aggregates.
Biochem Biophys Rep, 2, 2015
|
|
2J9H
 
 | | Crystal structure of human glutathione-S-transferase P1-1 cys-free mutant in complex with S-hexylglutathione at 2.4 A resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE P, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Hegazy, U.M, Hellman, U, Mannervik, B. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modulating Catalytic Activity by Unnatural Amino Acid Residues in a Gsh-Binding Loop of Gst P1-1.
J.Mol.Biol., 376, 2008
|
|
7NDZ
 
 | | ThyX reconstituted with N5-carbinolamine flavin | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7NDW
 
 | | ThyX-FADH2 soaked with 20 mM Formaldehyde | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
4ZK3
 
 | |
6VE5
 
 | |
6TXF
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | Descriptor: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | Authors: | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
7LUO
 
 | | N-terminus of Skp2 bound to Cyclin A | | Descriptor: | S-phase kinase-associated protein 2,Cyclin-A2, Skp2 Motif 1 uncharacterized fragment 1, Skp2 Motif 1 uncharacterized fragment 2 | | Authors: | Kelso, S, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Bipartite binding of the N terminus of Skp2 to cyclin A.
Structure, 29, 2021
|
|
6E2M
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-methyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2N
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl][3,4'-bipyridine]-2'-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2O
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | 4-(4-cyclopropyl-1H-imidazol-1-yl)-N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
8IBX
 
 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
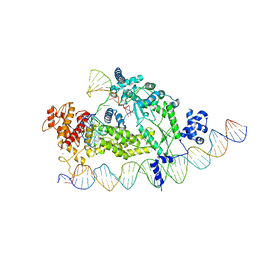 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
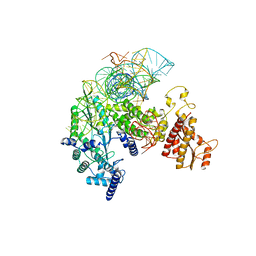 | | Structure of R2 with 5'ORF and 3'UTR | | Descriptor: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBY
 
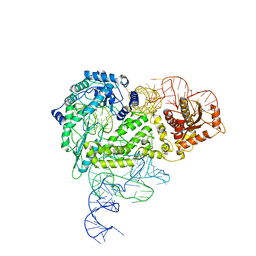 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3G
 
 | | Rd1NTF2_04 with long sheet | | Descriptor: | Rd1NTF2_04 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
