2F3A
 
 | |
2F3B
 
 | | Mechanism of displacement of a catalytically essential loop from the active site of fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, PHOSPHATE ION, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-11-20 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
2F3C
 
 | | Crystal structure of infestin 1, a Kazal-type serineprotease inhibitor, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Campos, I.T.N, Tanaka, A.S, Barbosa, J.A.R.G. | | Deposit date: | 2005-11-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Kazal-type inhibitors infestins 1 and 4 differ in specificity but are similar in three-dimensional structure.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2F3D
 
 | | Mechanism of displacement of a catalytically essential loop from the active site of fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
2F3E
 
 | | Crystal Structure of the Bace complex with AXQ093, a macrocyclic inhibitor | | Descriptor: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Rondeau, J.-M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease beta-site amyloid precursor protein cleaving enzyme (BACE).
J.Med.Chem., 49, 2006
|
|
2F3F
 
 | | Crystal Structure of the Bace complex with BDF488, a macrocyclic inhibitor | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease beta-site amyloid precursor protein cleaving enzyme (BACE).
J.Med.Chem., 49, 2006
|
|
2F3G
 
 | | IIAGLC CRYSTAL FORM III | | Descriptor: | GLUCOSE-SPECIFIC PHOSPHOCARRIER | | Authors: | Feese, M, Comolli, L, Meadow, N, Roseman, S, Remington, S.J. | | Deposit date: | 1997-10-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural studies of the Escherichia coli signal transducing protein IIAGlc: implications for target recognition.
Biochemistry, 36, 1997
|
|
2F3I
 
 | | Solution Structure of a Subunit of RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerases I, II, and III 17.1 kDa polypeptide | | Authors: | Kang, X, Jin, C. | | Deposit date: | 2005-11-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, biochemical, and dynamic characterizations of the hRPB8 subunit of human RNA polymerases
J.Biol.Chem., 281, 2006
|
|
2F3J
 
 | |
2F3K
 
 | | Substrate envelope and drug resistance: crystal structure of r01 in complex with wild-type hiv-1 protease | | Descriptor: | (3S,4AS,8AS)-N-(TERT-BUTYL)-2-[(3S)-3-({3-(METHYLSULFONYL)-N-[(PYRIDIN-3-YLOXY)ACETYL]-L-VALYL}AMINO)-2-OXO-4-PHENYLBUTYL]DECAHYDROISOQUINOLINE-3-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N.M, Nalivaika, E.A, Heilek-Snyder, G, Cammack, N, Schiffer, C.A. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease.
Antimicrob.Agents Chemother., 50, 2006
|
|
2F3L
 
 | |
2F3M
 
 | | Structure of human GLUTATHIONE S-TRANSFERASE M1A-1A complexed with 1-(S-(GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXADIENATE ANION | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
2F3N
 
 | |
2F3O
 
 | |
2F3P
 
 | |
2F3Q
 
 | |
2F3R
 
 | |
2F3S
 
 | |
2F3T
 
 | |
2F3U
 
 | |
2F3V
 
 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F3W
 
 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F3X
 
 | |
2F3Y
 
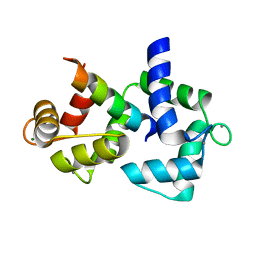 | | Calmodulin/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
2F3Z
 
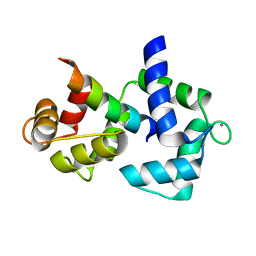 | | Calmodulin/IQ-AA domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent L-type calcium channel alpha-1C subunit | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
