7TS7
 
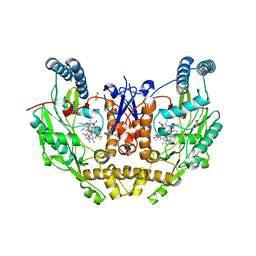 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-methyl-6-(3-(methylamino)propyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-[3-(methylamino)propyl]pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-Aminopyridines with a shortened amino sidechain as potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem., 69, 2022
|
|
9CW8
 
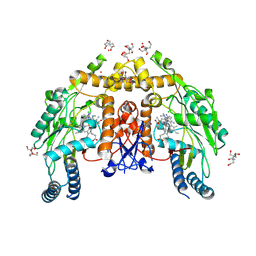 | | Structure of human endothelial nitric oxide synthase heme domain bound with 3-(6-amino-4-methylpyridin-2-yl)-5-((methylamino)methyl)benzonitrile dihydrochloride | | Descriptor: | (3P)-3-(6-amino-4-methylpyridin-2-yl)-5-[(methylamino)methyl]benzonitrile, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Truncated pyridinylbenzylamines: Potent, selective, and highly membrane permeable inhibitors of human neuronal nitric oxide synthase.
Bioorg.Med.Chem., 124, 2025
|
|
9CW4
 
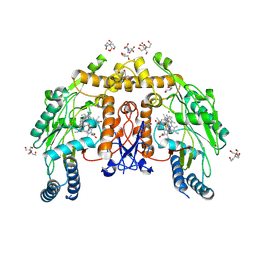 | | Structure of human endothelial nitric oxide synthase heme domain bound with 6-(3-fluoro-5-((methylamino)methyl)phenyl)-4-methylpyridin-2-amine dihydrochloride | | Descriptor: | (6M)-6-{3-fluoro-5-[(methylamino)methyl]phenyl}-4-methylpyridin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Truncated pyridinylbenzylamines: Potent, selective, and highly membrane permeable inhibitors of human neuronal nitric oxide synthase.
Bioorg.Med.Chem., 124, 2025
|
|
9CW6
 
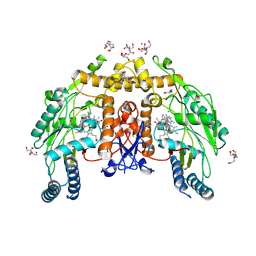 | | Structure of human endothelial nitric oxide synthase heme domain bound with 6-(3-chloro-5-((dimethylamino)methyl)phenyl)-4-methylpyridin-2-amine dihydrochloride | | Descriptor: | (6P)-6-{3-chloro-5-[(dimethylamino)methyl]phenyl}-4-methylpyridin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Truncated pyridinylbenzylamines: Potent, selective, and highly membrane permeable inhibitors of human neuronal nitric oxide synthase.
Bioorg.Med.Chem., 124, 2025
|
|
7B97
 
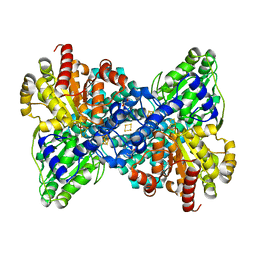 | | CooS-V with oxidized hybrid cluster by hydroxylamine for 30 min | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 3,5-dioxa-7-thia-1-thionia-2$l^{2},4$l^{2},6$l^{3},8$l^{2}-tetraferrabicyclo[4.2.0]octane, Carbon monoxide dehydrogenase, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Morphing [4Fe-3S-nO]-Cluster within a Carbon Monoxide Dehydrogenase Scaffold.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8U5I
 
 | | Crystal Structure of human IDO1 bound to Compound 23 | | Descriptor: | (1R,2S)-8-(ethanesulfonyl)-2-[(4R,5S,9aM)-5H-imidazo[5,1-a]isoindol-5-yl]-8-azaspiro[4.5]decan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Steinbacher, S, Lammens, A, Harris, S.F. | | Deposit date: | 2023-09-12 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploiting Stereo-Complexity to Identify Spirocyclic Imidazoisoindoles as Potent and Orally Bioavailable Dual Inhibitors of Indoleamine-2,3-Dioxygenase and Tryptophan-2,3-Dioxygenase
To Be Published
|
|
9GM3
 
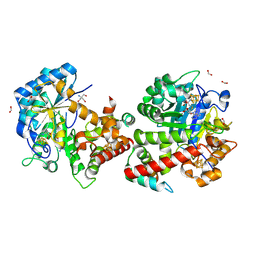 | | Crystal structure of the complex formed between the radical SAM protein ChlB and the leader region of its precursor substrate ChlA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ChlA, ChlB radical SAM domain, ... | | Authors: | de la Mora, E, Ruel, J, Usclat, A, Martin, L, Amara, P, Morinaka, B, Nicolet, Y. | | Deposit date: | 2024-08-28 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Peptide Recognition and Mechanism of the Radical S -Adenosyl-l-methionine Multiple Cyclophane Synthase ChlB.
J.Am.Chem.Soc., 147, 2025
|
|
5MOG
 
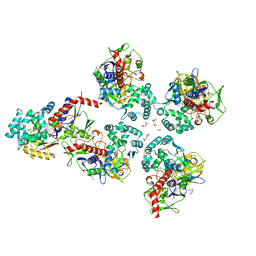 | | Oryza sativa phytoene desaturase inhibited by norflurazon | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brausemann, A, Gemmecker, S, Koschmieder, J, Beyer, P, Einsle, O. | | Deposit date: | 2016-12-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation.
Structure, 25, 2017
|
|
6RTP
 
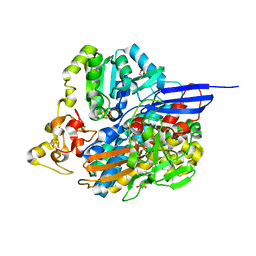 | |
6H0Y
 
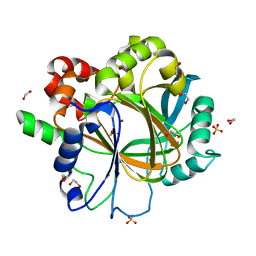 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022 | | Descriptor: | (2~{R})-3-(4-methoxyphenyl)-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.212 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022
To be published
|
|
7US8
 
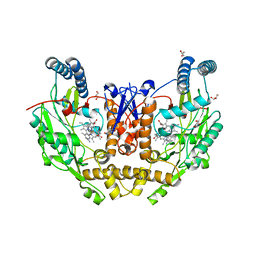 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(4-(dimethylamino)butyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[4-(dimethylamino)butyl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 2-Aminopyridines with a shortened amino sidechain as potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem., 69, 2022
|
|
8VHL
 
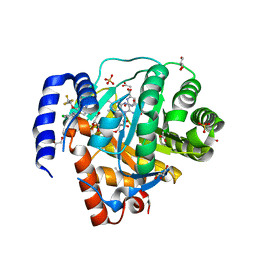 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
6E87
 
 | |
9ENK
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with L-phenylalanine | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, L-amino acid oxidase 4, PHENYLALANINE, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 598, 2024
|
|
5ZFB
 
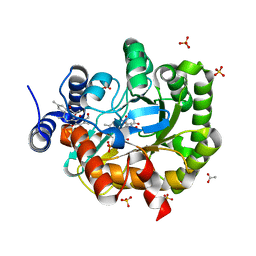 | | Structure of human dihydroorotate dehydrogenase in complex with ascofuranone (open-form) | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2Z,6Z,8E)-11-hydroxy-3,7,11-trimethyl-10-oxododeca-2,6,8-trien-1-yl]-2-methylbenzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
5O4Q
 
 | |
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5LBQ
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N2-(3-(dimethylamino)propyl)-6,7-dimethoxy-N4,N4-dimethylquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
7V0D
 
 | | Crystal structure of halogenase CtcP from Kitasatospora aureofaciens | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hou, C, Tsodikov, O.V. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and complex formation of halogenase CtcP and FAD reductase CtcQ from the chlortetracycline biosynthetic pathway
To Be Published
|
|
6G5W
 
 | | Crystal Structure of KDM4A with compound YP-03-038 | | Descriptor: | (4~{R})-5-methyl-4-phenyl-2-pyridin-2-yl-pyrazolidin-3-one, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
7Q5X
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH 2-OXOGLUTARATE (2OG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
7Q5V
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
5G3U
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with its inhibitor 2-(1H-indol-3-ylmethyl)prop-2- enoic acid | | Descriptor: | 2-[(1H-indol-3-yl)methyl]prop-2-enoic acid, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Biosynthesis of Violacein, Structure and Function of l-Tryptophan Oxidase VioA from Chromobacterium violaceum.
J.Biol.Chem., 291, 2016
|
|
6S07
 
 | | Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen. | | Descriptor: | Abz-ALA-THR-THR-PRO-LEU-CYS-GLY-PRO-SER-ARG-ALA-SER-ILE-LEU-SER-GLY-ARG, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Leisinger, F, Miarzlou, D.A, Seebeck, F.P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Chem Sci, 10, 2019
|
|
