7OVU
 
 | |
7OVV
 
 | | Crystal structure of the Arabidopsis thaliana thialysine acetyltransferase AtNATA2 | | Descriptor: | Probable acetyltransferase NATA1-like, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Layer, D, Kopp, J, Sinning, I. | | Deposit date: | 2021-06-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the Arabidopsis thaliana thialysine acetyltransferase AtNATA2
To Be Published
|
|
7A10
 
 | | LppS with covalent adduct derived from 1g | | Descriptor: | 4-methoxycyclohexa-2,5-diene-1-thione, L,D-transpeptidase 2 | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A1C
 
 | |
7A1E
 
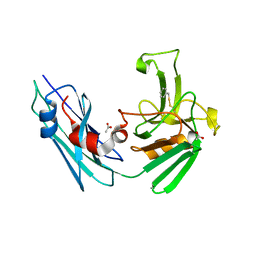 | | LppS with covalent adduct derived from 1c | | Descriptor: | ACETATE ION, L,D-transpeptidase 2, phenylmethanethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A11
 
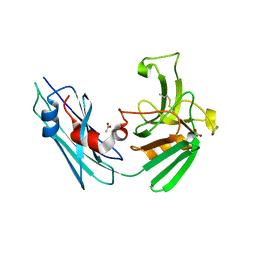 | | LppS with covalent adduct derived from 1E | | Descriptor: | ACETATE ION, L,D-transpeptidase 2, propane-1-thiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
6ET5
 
 | | Reaction centre light harvesting complex 1 from Blc. virids | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Qian, P, Siebert, C.A, Canniffe, D.P, Wang, P, Hunter, C.N. | | Deposit date: | 2017-10-25 | | Release date: | 2018-04-11 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Blastochloris viridis LH1-RC complex at 2.9 angstrom.
Nature, 556, 2018
|
|
7A0Z
 
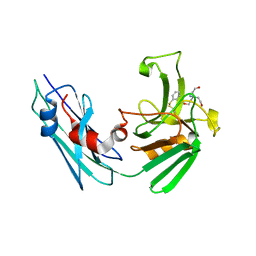 | | LppS with covalent adduct derived from 1b | | Descriptor: | L,D-transpeptidase 2, TRIS(HYDROXYETHYL)AMINOMETHANE, benzenethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
6ZXO
 
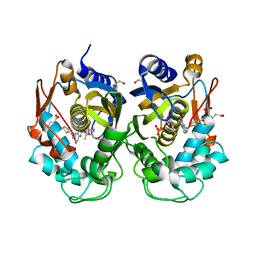 | |
8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
7PCY
 
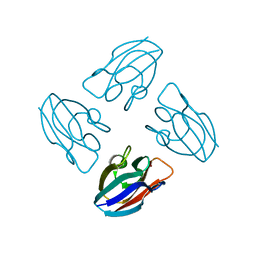 | | THE CRYSTAL STRUCTURE OF PLASTOCYANIN FROM A GREEN ALGA, ENTEROMORPHA PROLIFERA | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Collyer, C.A, Guss, J.M, Freeman, H.C. | | Deposit date: | 1989-09-22 | | Release date: | 1990-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of plastocyanin from a green alga, Enteromorpha prolifera.
J.Mol.Biol., 211, 1990
|
|
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
6ZRX
 
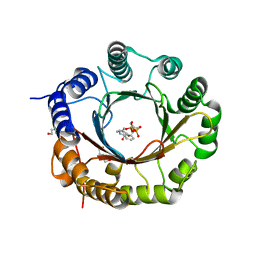 | | Crystal structure of 6-dimethylallyltryptophan synthase from Micromonospora olivasterospora in complex with DMASPP and Trp | | Descriptor: | DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase, S-(3-methylbut-2-en-1-yl) trihydrogen thiodiphosphate, ... | | Authors: | Ostertag, E, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
1DLV
 
 | |
5MX2
 
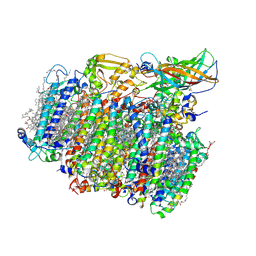 | | Photosystem II depleted of the Mn4CaO5 cluster at 2.55 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zhang, M, Bommer, M, Chatterjee, R, Hussain, R, Kern, J, Yano, J, Dau, H, Dobbek, H, Zouni, A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural insights into the light-driven auto-assembly process of the water-oxidizing Mn4CaO5-cluster in photosystem II.
Elife, 6, 2017
|
|
8B64
 
 | | Cryo-EM structure of RC-LH1-PufX photosynthetic core complex from Rba. capsulatus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.589 Å) | | Cite: | Cryo-EM structure of a monomeric RC-LH1-PufX supercomplex with high-carotenoid content from Rhodobacter capsulatus.
Structure, 31, 2023
|
|
7PPS
 
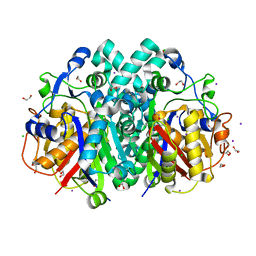 | | apo FabB from Pseudomonas aeruginosa with single point mutation C161A | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 1, CHLORIDE ION, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V. | | Deposit date: | 2021-09-15 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa FabB C161A, a template for structure-based design for new antibiotics.
F1000Res, 10, 2021
|
|
7PIL
 
 | | Cryo-EM structure of the Rhodobacter sphaeroides RC-LH1-PufXY monomer complex at 2.5 A | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the monomeric Rhodobacter sphaeroides RC-LH1 core complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
8BIP
 
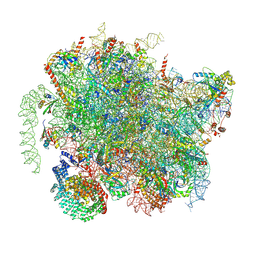 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BJQ
 
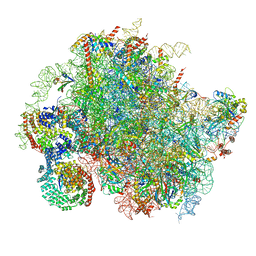 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
1E14
 
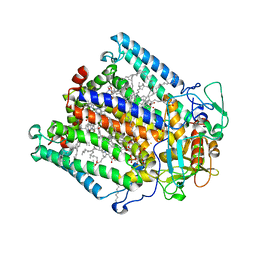 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH PHE M197 REPLACED WITH ARG (CHAIN M, FM197R) AND GLY M203 REPLACED WITH ASP (CHAIN M, GM203D) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Ridge, J.P, McAuley, K.E, Cogdell, R.J, Isaacs, N.W, Jones, M.R. | | Deposit date: | 2000-04-18 | | Release date: | 2000-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Consequences of the Replacement of Glycine M203 with Aspartic Acid in the Reaction Center from Rhodobacter Sphaeroides.
Biochemistry, 39, 2000
|
|
6F4D
 
 | |
7PBW
 
 | | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Qian, P, Swainsbury, D.J.K, Croll, T.I, Castro-Hartmann, P, Sader, K, Divitini, G, Hunter, C.N. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Cryo-EM Structure of the Rhodobacter sphaeroides Light-Harvesting 2 Complex at 2.1 angstrom.
Biochemistry, 60, 2021
|
|
7PQD
 
 | | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 A: the structural basis for dimerisation | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 angstrom : the structural basis for dimerisation.
Biochem.J., 478, 2021
|
|
