5WBF
 
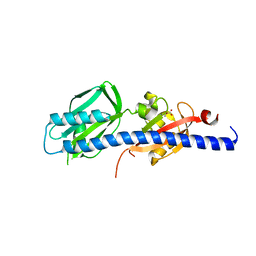 | | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | | Descriptor: | GLYCEROL, LACTIC ACID, Methyl-accepting chemotaxis transducer (TlpC) | | Authors: | Machuca, M.A, Johnson, K.S, Liu, Y.C, Steer, D.L, Ottemann, K.M, Roujeinikova, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate.
Sci Rep, 7, 2017
|
|
5WB6
 
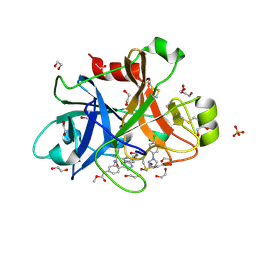 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5WBC
 
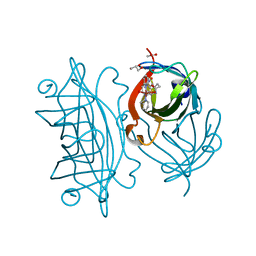 | | Designed Artificial Cupredoxins - WT | | Descriptor: | ACETATE ION, GLYCEROL, Streptavidin, ... | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Modular Artificial Cupredoxins.
J. Am. Chem. Soc., 138, 2016
|
|
5WBP
 
 | |
5WC6
 
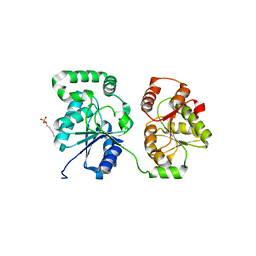 | |
5WCF
 
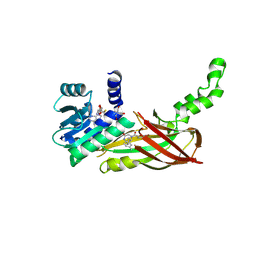 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | (5R)-4-(5-bromofuran-2-carbonyl)-5-(4-fluorophenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Zeng, H, Hutch, A, Seitova, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with MTLLE1441
to be published
|
|
5WC8
 
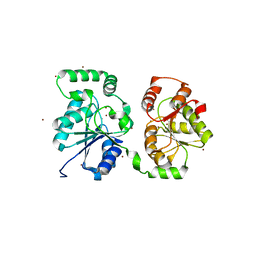 | |
4NH4
 
 | | Structure of the binary complex of a zingiber officinale double bond reductase in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Zingiber officinale double bond reductase | | Authors: | Langlois D'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2013-11-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of zingiber officinale double bond reductase
To be Published
|
|
4NHO
 
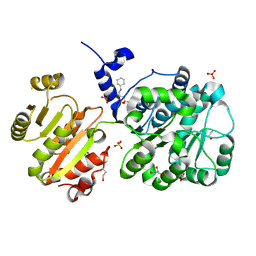 | | Structure of the spliceosomal DEAD-box protein Prp28 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Moehlmann, S, Neumann, P, Ficner, R. | | Deposit date: | 2013-11-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NI5
 
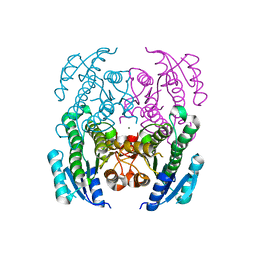 | | Crystal Structure of a Short Chain Dehydrogenase from Brucella suis | | Descriptor: | MAGNESIUM ION, Oxidoreductase, short-chain dehydrogenase/reductase family protein | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Short Chain Dehydrogenase from Brucella suis
TO BE PUBLISHED
|
|
5WDC
 
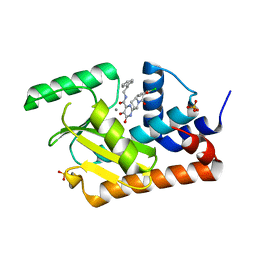 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9e (SRI-29843) | | Descriptor: | 2-{(2S)-1-[(2-chlorophenoxy)acetyl]pyrrolidin-2-yl}-5-hydroxy-6-oxo-N-(2-phenylethyl)-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-07-04 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5WDI
 
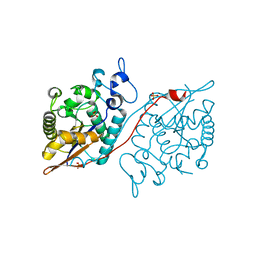 | | Structure of Human Sts-2 histidine phosphatase domain | | Descriptor: | SULFATE ION, Ubiquitin-associated and SH3 domain-containing protein A | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
5WE8
 
 | |
4NL9
 
 | |
5WE7
 
 | |
4NLU
 
 | | Poliovirus Polymerase - G289A Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NM9
 
 | |
4NCL
 
 | |
5WJD
 
 | | Crystal structure of Naa80 bound to acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, CG8481, isoform B, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4NF2
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis in complex with carbamoyl phosphate and L-norvaline | | Descriptor: | CHLORIDE ION, NORVALINE, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Handing, K, Cymborowski, M.T, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
5WJJ
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, N-{4-[2-(4-fluoro-3-methylphenyl)imidazo[1,2-b]pyridazin-3-yl]pyridin-2-yl}-2-methyl-1-oxo-1lambda~5~-pyridine-4-carboxamide | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-07-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
4NFN
 
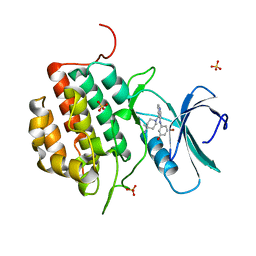 | | Human tau tubulin kinase 1 (TTBK1) complexed with 3-({5-[(4-amino-4-methylpiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-bromophenol | | Descriptor: | 3-({5-[(4-amino-4-methylpiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-bromophenol, SULFATE ION, Tau-tubulin kinase 1 | | Authors: | Sheriff, S. | | Deposit date: | 2013-10-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The structure of human tau-tubulin kinase 1 both in the apo form and in complex with an inhibitor.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5WMP
 
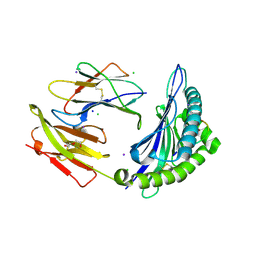 | | Crystal Structure of HLA-B7 in complex with TPR, a CMV peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inability To Detect Cross-Reactive Memory T Cells Challenges the Frequency of Heterologous Immunity among Common Viruses.
J. Immunol., 200, 2018
|
|
5WM2
 
 | |
4NGG
 
 | |
