5IMR
 
 | | Structure of ribosome bound to cofactor at 5.7 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5IB7
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet, near-cognate tRNALys with U-G mismatch in the A-site and antibiotic paromomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
5IMQ
 
 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5IT9
 
 | | Structure of the yeast Kluyveromyces lactis small ribosomal subunit in complex with the cricket paralysis virus IRES. | | Descriptor: | 18S ribosomal RNA, Cricket paralysis virus IRES RNA, MAGNESIUM ION, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
5IT8
 
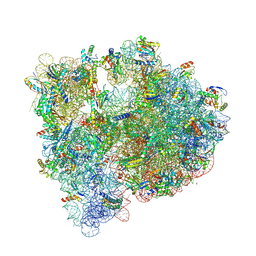 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IT7
 
 | | Structure of the Kluyveromyces lactis 80S ribosome in complex with the cricket paralysis virus IRES and eEF2 | | Descriptor: | (2S)-1-amino-N,N,N-trimethyl-1-oxobutan-2-aminium, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
7JQB
 
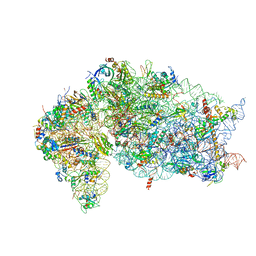 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JSS
 
 | |
7JQL
 
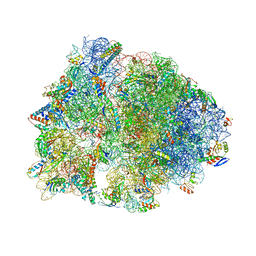 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-001, mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
7JT3
 
 | |
7JSW
 
 | |
7JQM
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-002, mRNA, and deacylated P-site tRNA at 3.05A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K53
 
 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K54
 
 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K52
 
 | | Near post-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7JIL
 
 | | 70S ribosome Flavobacterium johnsoniae | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Ortega, J. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of sequestration of the anti-Shine-Dalgarno sequence in the Bacteroidetes ribosome.
Nucleic Acids Res., 49, 2021
|
|
7JT2
 
 | |
7K00
 
 | | Structure of the Bacterial Ribosome at 2 Angstrom Resolution | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Watson, Z.L, Ward, F.R, Meheust, R, Ad, O, Schepartz, A, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2020-09-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structure of the bacterial ribosome at 2 angstrom resolution.
Elife, 9, 2020
|
|
7JT1
 
 | |
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7KGB
 
 | |
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K55
 
 | | Near post-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure III-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7JSZ
 
 | |
