7Z5J
 
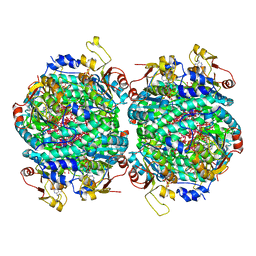 | | The molybdenum storage protein loaded with tungstate | | Descriptor: | 1,1,3,3,5,7,7,9,11,15,15-undecakis($l^{1}-oxidanyl)-2$l^{4},4$l^{3},6$l^{5},8,10,12,14,16,17,18,19$l^{3},20,21,22,23-pentadecaoxa-1$l^{6},3$l^{6},5$l^{6},7$l^{6},9$l^{6},11$l^{6},13$l^{6},15$l^{6}-octatungstapentadecacyclo[7.7.1.1^{1,13}.1^{3,5}.1^{3,15}.1^{5,7}.1^{5,11}.1^{7,11}.0^{2,13}.0^{2,15}.0^{4,13}.0^{6,9}.0^{6,11}.0^{6,13}.0^{9,19}]tricosane, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ermler, U, Aziz, I, Kaltwasser, S, Kayastha, K, Vonck, J. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates.
J.Inorg.Biochem., 234, 2022
|
|
5WYK
 
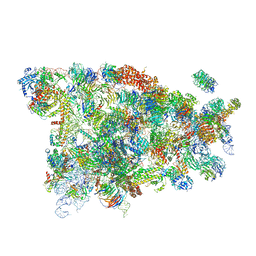 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2017-05-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
7YQT
 
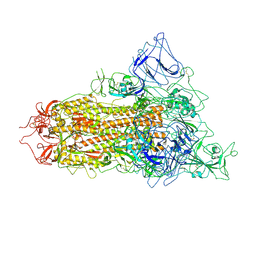 | | SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR3
 
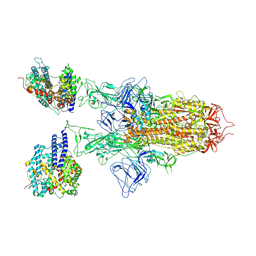 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQV
 
 | | pH 5.5 SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR2
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQU
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR1
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with XG2v024 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQZ
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7ZCU
 
 | | PucA-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCE
 
 | |
7YQX
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQY
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQW
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
1YAR
 
 | | Structure of Archeabacterial 20S proteasome mutant D9S- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAU
 
 | | Structure of Archeabacterial 20S proteasome- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
7YP3
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with elaiophylin | | Descriptor: | ACETATE ION, Elaiophylin, GLYCEROL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Z8F
 
 | |
7ZAG
 
 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA,mRNA, aIF1A and the C-terminal domain of aIF5B. | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
5WQ1
 
 | |
7ZAH
 
 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA, aIF1A and aIF5B | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
5X51
 
 | | RNA Polymerase II from Komagataella Pastoris (Type-3 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.996 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
1Y82
 
 | | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Horanyi, P, Tempel, W, Habel, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus
To be published
|
|
2AJ8
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
2AI0
 
 | | Anti-Cocaine Antibody 7.5.21, Crystal Form III | | Descriptor: | GLYCEROL, Immunoglobulin Heavy Chain, Immunoglobulin Light Chain kappa, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of Packing: Four Crystal Forms of an Anti-Cocaine Antibody 7.5.21
To be Published
|
|
